Proteomic analysis of a copper mine isolated fungus Rhizopus microspores IOC 4686 when exposed to copper sulfate
1Dempster MS Lab, Chemical Engineering Department of Polytechnic School of the University of São Paulo, Rua do Lago, 250, Bloco B 3 andar. São Paulo-SP, 05508-080, Brazil
2Federal University of São Paulo, Rua Tres de Maio, 100, 2 andar. São Paulo-SP, 05508-080, Brazil
Author and article information
Cite this as
Dias M, Gomes de Lacerda JTJ, Andrade LM, Oller do Nascimento CA, Mendes MA, et al. (2022) Proteomic analysis of a copper mine isolated fungus Rhizopus microspores IOC 4686 when exposed to copper sulfate. Arch Biochem. 2022; 5(1): 001-010. Available from: 10.17352/ab.000005
Copyright License
© 2022 Dias M, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.The fungus Rhizopus microsporus, known for its absorption capacity for toxic metals was used to understand the green metal recovery via analysis of its physiology under metal stress conditions through proteomic methods. To investigate the effects of copper stress on fungus, R. microsporus IOC 4686, isolated from the mine environment, was exposed to copper ions (50mgL-1) for 48 h. This study was performed only on copper exposure. Tryptic and chymotryptic extracts of proteins were analyzed by nano- liquid chromatography-tandem mass spectrometry, identification was performed by PEAKS Studio 8.5. Proteins were classified according to their molecular function and biological process. Enzymes, such as catalase, superoxide dismutase, and cytochrome c peroxidase were found in the presence and absence of copper ions. However, only in presence of copper ions, was observed the presence of heat shock proteins (HPS 20, HPS 70, and HPS 78) and metalloproteins (GrpE protein homolog and cytochrome P450). These classes of proteins have been produced by cells in response to stress conditions. The control group (absence of copper ions) also presented antioxidant enzymes suggesting that the fungus isolated from the mine environment already has adapted to the copper. The presence of these proteins suggested a physiological response of R. microsporus IOC 4686 to oxidative stress induced by copper.
Oxidative stress is the unbalance in the regulatory system that causes physiological alteration of microorganisms to compensate for the changing environment by generating molecules to neutralize radical species through oxygen metabolism [1]. The oxidative stress of microorganisms has aroused great interest due to the relationship between the physiological responses and the damage suffered by cellular metabolism.
Toxic metals, when exceeding the resistance capacity of organisms, induce physiological damage that interferes in biological processes, such as enzymatic activity, DNA transcription, and translation or cell integrity. In general, metals such as copper and chromium, despite having an important role in biochemistry, can be toxic at high concentrations [2].
The expansion of mining processes has quantitatively increased the complexity of toxic waste released into the environment. In this sense, the extraction of copper, mainly in Brazil, increased by 90% between 1998 and 2015 due to primary and secondary copper production. Thus, it was estimated that in 2030, national metal production will reach up to 374,000 tons/year, and Brazilian reserves were estimated at 21 million tons [3]. Screening of processes to reduce the environmental damage produced by toxic metals has intensified in recent years. Some studies have investigated the application of microorganisms, such as bacteria, microalgae, and fungi, in bioremediation processes, which have been adapted to reduce toxic metal concentrations [4,5]. In general, compared with conventional methods such as precipitation, reverse osmosis, or ion exchange, 50-65% of the costs could be saved by applying bioremediation to treat the soil Pb- polluted [2].
Filamentous fungi are a versatile group in bioremediation processes, as they can grow under extreme temperatures, nutrient availability, toxic metal conditions, and pH variations (Anand, et al. 2006). Filamentous fungi react differently when exposed to pollutants, producing upregulation or downregulation responses through organic compounds, mainly proteins, although their use requires a complete understanding of response mechanisms [6]. Protein response, such as heat shock and Superoxide dismutase is used as conventional biomarkers due to the well-characterized presence in oxidoreductive intracellular alterations [7]. Rhizopus is a fungus genus known for its ability to sequester and precipitate metals, producing intracellular and extracellular enzymes which can be used for metal recovery from contaminated soils [8,9].
Proteomics is one of the most recent techniques used in the identification of new biomarkers, which can be proteins or other cellular components that react specifically to the presence of a pollutant, indicating environmental damage [10]. Therefore, proteomics approaches provide remarkable information on protein expression in different environments (e.g., polluted and unpolluted), enabling the use of proteins as biomarkers [7,10]. The present work studied the proteomic profile from R. microsporus strain IOC 4686 grown in the presence of copper. NanoLC-MS/MS system was used to identify proteins Cu2+response. This study aimed to use proteomic profiles to better understand of bioremediation process and improve the in-situ bioremediation.
Materials and methods
R. microsporus fungus
R. microsporus strain IOC 4686 was isolated from a mine environment (local copper concentration of about 80mgkg 1) located in Pará, Brazil. Fungal identification was performed using the MALDI-TOF Score: 2.216) BioTyper 3.1 database (Bruker Daltonics, Germany) [11]. Isolated fungal strains were morphologically identified by FioCruz Institute and kept in its filamentous fungus collection.
Fungal growth and inhibition
The fungus was inoculated and maintained in Potato Dextrose Agar (PDA), Sigma Aldrich solution (42gL-1). The inoculum was performed using the spores collected after 10 days at 25°C, from solid culture. The spores (6.7×106/ mL-) were suspended in glycerol solution (20% v/v), and stored at -80°C [4]. These inoculums were used in the fungal growth and inhibitory concentration tests.
Fungal growth inhibition was determined by exposing R. microsporus IOC 4686 to a PDB medium containing copper ions (5-120mgL-1) [12,13], at 30°C for 80h under 150rpm..
According to the research done by GlobalData, results have shown that IL-6 that acts as a biomarker for increased immune response and inflammation, is more evident in patients who were suffering from pneumonia. All but two of the highlighted clinical trials done by the GlobalData Clinical Trials Database have shown results used biomarkers. And all of the trials which have achieved their endpoints used biomarkers. With the increase of COVID-19 global threat and the consecutive trials performed, it is a given that these trials will include biomarkers to help in trial advancement, assisting in patient diagnosis and selection of drug administration [2].
Other research studies that were conducted have found a noteworthy association between thrombocytopenia, lymphocytopenia, and the elevated levels of CRP, PCT, LDH, D-dimer and COVID-19 severity. The results have shown that it is inherently possible to be used as an early biomarker to improve the COVID-19 patient care, by detecting high-risk patients and providing fitting healthcare resources during the pandemic [3].
Biomass was determined by sampling 5ml every 8h, filtering in 0.45-μm pore membranes (Merck Millipore). After filtration, the membranes were microwaved at 228W for 15min and weighed to obtain the fungal mass [14]. The weight obtained for each sampling was used to calculate the growth rate. It was plotted the time of reaction by the weight of biomass.
To determine stress induced by copper, the fungus was grown in a PDB medium in the absence (control group) and presence of 50mgL-1 of copper ions (treated group). The experiments were carried out in triplicate.
Determination of copper biosorption
To determine metal ion absorption, the supernatants of cultures were collected after 48 and 60 h of growth in a medium containing 50mgL-1 of copper ions. Elemental composition was determined directly by the EDX instrument (Epsilon 3-XL-PANalytical) operated in 50kV, 99µA, standard detector, air drag medium configuration. Samples and standards were dispensed in 32 mm diameter vessels fitted with a 6µm polypropylene (PP) film.
Proteomic analysis
Fungus biomass was collected and lyophilized for protein extraction. Proteins were extracted by cell disruption by adding 50 µL of 70% formic acid (MS grade) to each sample (0.2mg) and shaken for 2min. Then, 50µL of acetonitrile was added, and samples were sonicated (Ultronique-Q30140A) for 15min at room temperature. The extracts were centrifuged (Mini Spin-Eppendorf) at 17,400×g for 30min at 6°C, and supernatants were collected for protein quantification. Protein concentrations were determined by measuring the absorbance at 595nm in a spectrophotometer (UV2600, Shimadzu) using the Bradford assay (Sigma-Aldrich) with bovine serum albumin as standard [15].
Identification of proteins by nanoLC-tandem mass spectrometry
Protein extract samples (2mg) were digested in a 1:50 (enzyme: substrate) ratio using two enzymes, trypsin (Sigma-Aldrich) and chymotrypsin (Roth). Trypsin was prepared in 400µL of NH4HCO3 solution (50mM) to reach a final concentration of 0.05µg µL-1, according to the manufacturer’s instructions. Subsequently, 5µL of the trypsin solution was added to each sample and incubated at 37°C for 24h. Chymotrypsin digestion of protein was performed by adding 50 µL of enzyme solution to each sample, incubating at 25°C for 24h. The digestion was stopped by adding 10μL of 10% (v/v) trifluoroacetic acid (TFA- Sigma-Aldrich) solution for 90min, at 37°C and 30°C for trypsin for chymotrypsin, respectively.
The peptides solution of each digestion was analyzed separately, using a nanoLC System (Thermo Fisher Scientific), containing a PepMap column (15cm×75µm; Thermo Scientific) by using a gradient from 2 to 98% (v/v) acetonitrile with 0.1% TFA for 180 min [5,16] and ESI-Q-TOF mass spectrometry (Impact II mass spectrometer Bruker Daltonics) The nanoLC-ESI-Q-TOF system was operated in extracted ion mode, and chromatograms and full- scan MS spectra were acquired at a rate of 0.5Hz. MS precursors and MS/MS product ions were acquired over a 50-3000 m/z range, and the collision-induced dissociation energy ranged from 7 to 70eV.
Database searching and statistical analysis
Data file (.d) analysis was performed in PEAKS Studio 8.5 software (Bioinformatics Solutions Inc., Waterloo, Canada), and MS/MS spectra were submitted for de novo analysis and database search using peaks B, PTM, and Spider tools [17]. Considering that R. microsporus has a low number of curated proteins in the database, de novo sequenced peptides with average local confidence scores ≥ 50% were compared with the R. microsporus UniProt/TrEMBL database (53,362 sequences, downloaded in January 2021). Parameters used were precursor mass tolerance of 20 ppm; fragment mass tolerance at 0.025 Da; trypsin or chymotrypsin was set as the specific enzymes and up to two missed cleavages were required; carbamidomethylation (Cys) as fixed modification and oxidation (Met) set as variable modifications, with maximal 3 modifications per peptide in SPIDER outcomes. The combination of multiple runs (trypsin and chymotrypsin) in a single project and a database search were performed for each experimental group. A false discovery rate (FDR) threshold of 1% on the peptide-spectrum match (PSM) and at least one unique peptide was applied to protein identifications. The UniProt classification system was used to analyze all the identified proteins by specifically selecting R. microsporus, followed by ontology relations, such as molecular function, and biological process.
Results
Effects of copper on the fungal growth profile
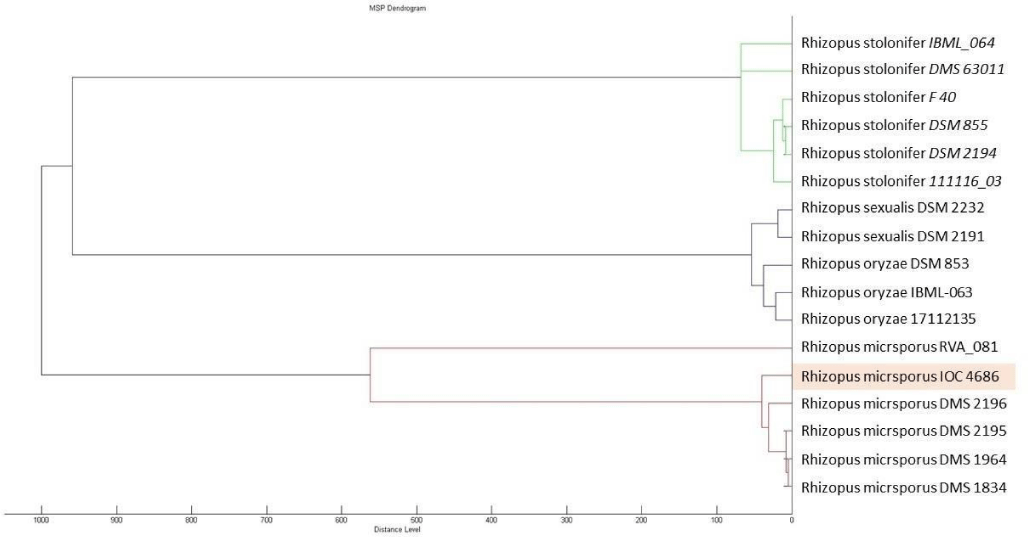
IOC 4686 was obtained from a collection of fungi isolated from the Sossego mine environment, during the screening of the copper recovery assay. Dendrogram (Figure 1) showed genetic relation between IOC 4686 strain and other Rhizopus species cataloged in the Biotyper database.
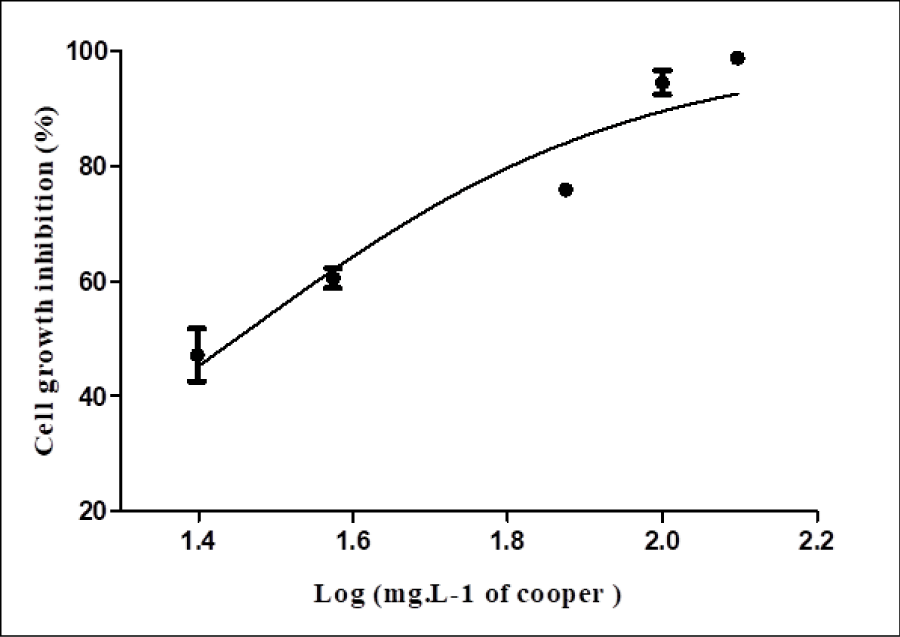
R. microsporus showed resistance to 50mgL-1 of copper ions, close to the IC50 value (28 mg L-1). The cell growth density of 4.66gL-1 when exposed to a concentration of 50mgL-1 copper ions (Figure 2), decreased to 0.11gL-1in the higher tested concentration (125mgL-1). Based on this, 50mgL-1 of copper ions was considered to induce a non-lethal stress level for proteomic study.
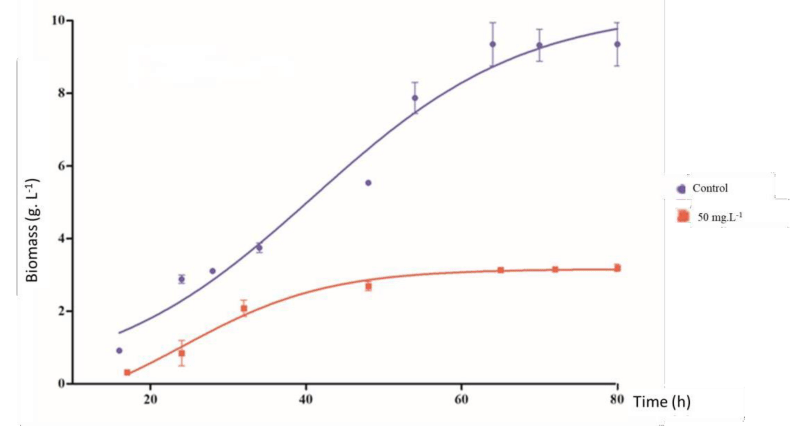
In absence of copper (control), the fungus showed a rapid adaptation phase, and accelerated growth (log phase) followed by a stationary phase occurred after approximately 80h. Cultures treated with copper showed a cell growth rate 65.14% lower than the control group (Figure 3).
Copper absorption
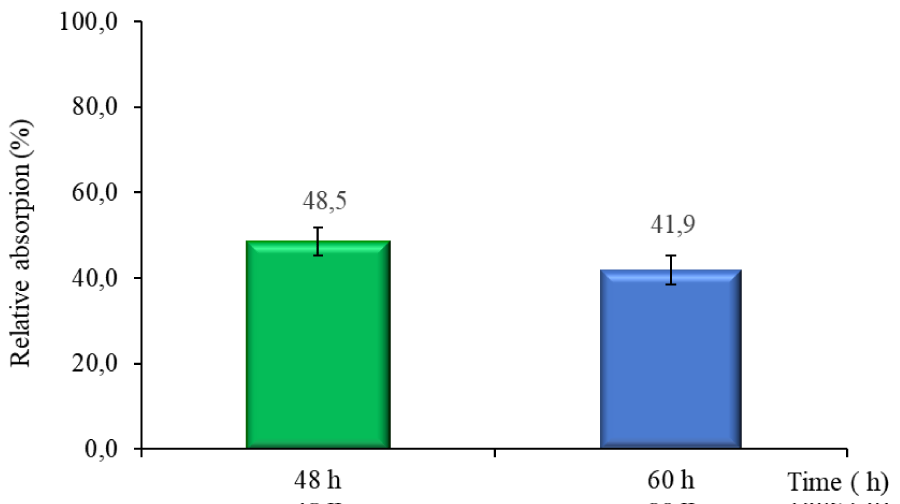
The results showed that the R. microsporus IOC 4686 fungus was able to remove metal from the medium culture. Figure 4 shows the copper concentration determined at two points of fungal growth by EDX analysis, indicating that two processes were occurring. During the first 48h copper absorption process, after 60h copper desorption process.
Cells exposed to 50mgL-1 of copper ions showed the highest copper absorption (48.6% of recovery) occurred after 48h of growth, then, after 60h (Figure 4), a metal ion desorption process was observed the fungus released 6,7% of metal. The lower copper concentration in the culture after 60 h of cultivation observed suggested that the fungus releases the absorbed copper to the medium.
Protein profile of R. microsporus IOC 4686 after copper exposure - Gene ontology (GO) functional enrichment.
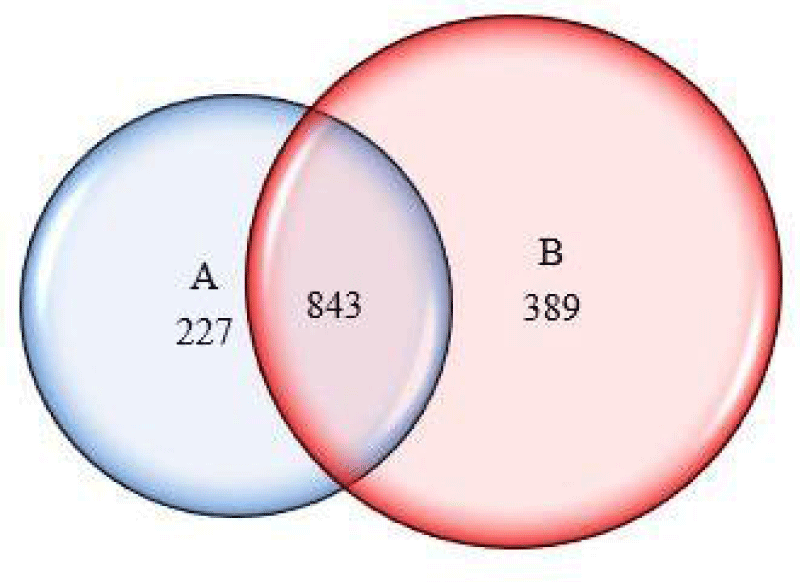
To compare the proteins profile in both groups (with and without copper), these proteins were assembled in a list. The protein set showed 1071 proteins in the control and 1232 proteins in the culture containing copper. The Venn diagram (Figure 5) showed that 843 proteins were common to both, mostly proteins related to the basal metabolism of the fungus.
Proteins involved in the oxidative process such as ATP-citrate (pro-S-)-lyase, pyruvate decarboxylase isozyme, Hsp71-like protein, glyceraldehyde-3-phosphate dehydrogenase, elongation factor 1-alpha fructose-bisphosphate aldolase and ATP syntha0se subunit alpha were observed in fungi treated (Table 1).
HSPs proteins were identified in both groups, however, the HSPs, such as G-alpha protein, HSP (7, 70, 78, 90, and SSB1), and Small COP II coat GTPase were identified only in the culture containing copper (Table 1). Considering only proteins identified under stress conditions induced by copper, could highlight anti-oxidative process proteins, increasing 27.3% transporter activity protein (Cytochrome b-c1 complex subunit Rieske, mitochondrial and V-type proton ATPase subunit G), and 17.6% binding activity proteins (Ribonucloprotein and Aconitate hydratase, mitochondrial).
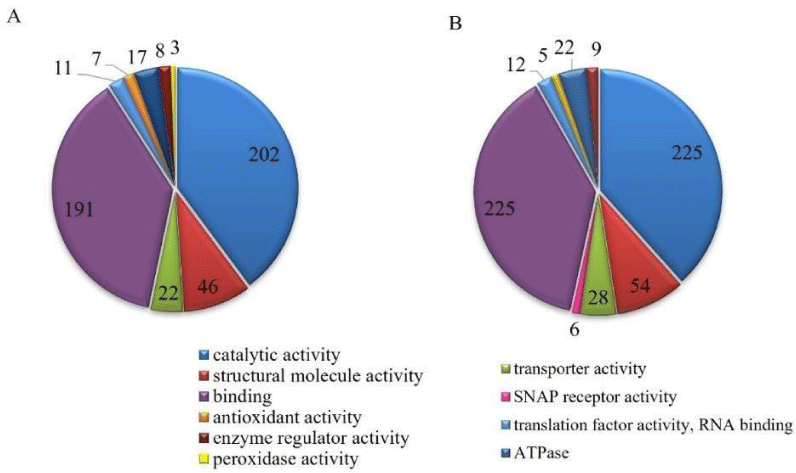
GO analysis (Figure 6) showed the presence of proteins belonging to molecular functions such as antioxidant, transporter, enzymatic regulator, translation factor (RNA binding), binding, structural molecular, ATPase, SNAP receptor, peroxidase, and catalytic. Proteins related to catalytic activity, binding, translation factors, structure, transporter, and enzyme regulator were found in both groups. In another way, SNAP receptor activity was present only in stress conditions.
The presence of selective stress-induced binding enzymes, such as calreticulin, cell division/GTP binding protein, tyrosine-tRNA ligase, Peptidase M18, amino-peptidase I, arginyl- tRNA synthetase, and ATP-dependent metallopeptidase Hfl, in both groups suggests that the fungi were adapted to stress environment.
In the copper-containing culture were observed an increase in the number of proteins related to transporter activity (27.3%), structural molecule, binding (17.5%), catalytic, translation factor (9.1%), and enzyme regulator (12.5%). Peroxidase function proteins were found only in the control group.
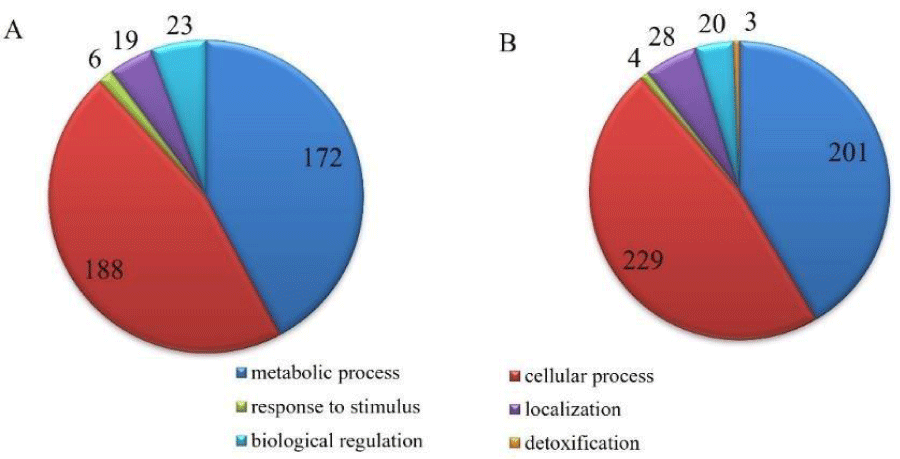
The different biological process of proteins is shown in Figure 7. Metabolic, cellular, biological regulation, response to stimulus, and localization process were observed in both groups, however, the copper treatment also presented a detoxification process, this is the only difference observed concerning the biological process.
In R. microsporus strain IOC 4686, the pyruvate dehydrogenase, and pyruvate kinase- carboxylase complex was identified in the copper-treated group.
Fungus under copper stress, showed an increased number of ATPase proteins isoforms, such as ATP-citrate (pro-S-)-lyase, ATP-dependent metallopeptidase Hfl, and C-terminal domain of alpha and beta subunits of F1 ATP synthase.
Induction of the expression of metalloproteins such as leukotriene A hydrolase, AFG3 family protein, and ATP-dependent metallopeptidase Hfl was observed in R. microsporus IOC 4686 in the presence of copper (Table 2).
Discussion
The results observed during the grown experiments showed that two processes occurred, the absorption and desorption processes. The second one occurred about 60h after, the fungus released 6.7% of copper ions recovered, this process occur probably due to the decrease of pH values caused by the metabolites secreted by the fungus [18]. Copper is an essential metal for several enzymatic and protein functions in the cell, contributing to the maintenance of homeostasis. However, when the limit of tolerance is exceeded, cell growth, metabolism, and protein expression can be modified [10,19,20].
The influence of copper ions on biological processes, such as blocking functional groups or denaturing enzymes, could explain the variation in R. microsporus IOC 4686 growth, mainly in the highest concentration of metal. Cell growth reduction has usually been used as a heavy metals tolerance indicator as well as in the determination of the metabolic effects on microorganisms [20]. To evaluate the metabolism of fungi under copper stress, samples were collected at the maximum copper absorption conditions (48h) to identify the proteins involved in the absorption and resistance to the metal.
Fungi expressed different proteins isoform in presence of copper, which suggested differences in cellular metabolism represented by alteration in enzymes involved in oxidative stress. Proteomic analysis of R. microsporus IOC 4686 identified proteins involved in basal metabolic pathways, such as carbohydrate consumption, protein biosynthesis, catabolism of secondary metabolites, energy production, and conversion. Mitochondrial pyruvate carrier and putative acetyl-CoA hydrolase/transferase, proteins responsible for changes in the tricarboxylic acid cycle (TCA) which are involved in the degradation of amino acids were observed in presence of copper.
To carry out the proteomics studies, we chose to use two different types of digestive enzymes: trypsin and chymotrypsin, since these enzymes breakdown in different amino acid residues, increasing the number of identified proteins, by generating different sets of peptides, improving protein sequence coverage helping the understood of the metabolism involved in the fungi metal recovery.
The results showed that the oxidative stress induced by copper in fungus isolated from the mine environment modified the protein profile. However, there is a lack of knowledge about the genes of R. microsporus involved in the absorption of copper in bioremediation processes, in the present work was established that proteins closely related to this process were induced by the presence of copper [21].
The proteomic analysis showed that some proteins related to copper response increased in several isoforms, and it was also observed the presence of proteins, that were not observed in the control (Table 1), these proteins were considered as possible biomarkers present in R. microspores which given to the fungus the ability to survive in a harmful environment. These proteins overexpression indicated modification in fungal metabolism as a response to the metal present in the medium.
Copper stress reduced the protein isoforms related to the mitochondrial respiration process in R. microsporus IOC 4686. Considering that ATP production is the main function of mitochondria [22], alteration in this function also causes alteration in the expression of NADH, FADH2, and cytochrome (P450, b5, c), since copper ions blocked the enzymes that compose the electron carrier chain. Proteins such as cytochrome P450, malic enzyme, acyl-CoA desaturase, among other proteins with oxidoreductase activity in the presence of NADPH were expressed in the presence of an oxidizing agent [23].
FMN-linked oxidoreductase (FMN) was expressed under copper stress. As the copper ions are blocking the electronic carriers the fungal cells must produce FMN to replace the blocked carrier. Additionally, FMN acts as the protein which maintains the electronic chain of cells, protecting the fungi of the osmotic lysis [10].
In the presence of copper, glycolytic pathway proteins were affected by metal, reducing the formation of ATP and its intermediates [24]. In contrast, proteins related to glycolysis and gluconeogenesis, such as pyruvate kinase and enolase, as well as glutamine synthetase and adenosylhomocysteinase, are proteins associated with the assimilation of copper by the cell, were identified in the absence and presence of copper ions. These proteins are essential to energetic metabolism, present in all organisms, their presence in the group under copper stress indicated that cooper ions inhibit the electronic chain, but they do not affect the ATP production, suggesting that isoforms stress-induced (present in the electronic chain) maintained cell energy production. The synthesis of isoforms allowed the production of pyruvate through the glycolytic pathway helping aerobic respiration activity. All these proteins play an important key role in R. microsporus strain IOC 4686 fungus homeostasis, being responsible to transport copper across the intracellular and cytoplasmic membrane using ATP [25].
Proteins related to the mitochondrial cytochrome b-c1 complex, which is activated by the Fe-S cofactor in the mitochondria, also participate in the transfer of electrons from ubiquinol to cytochrome during oxidative phosphorylation [22].
The presence of oxidizing agents in the mining environment, in which the fungus was isolated, induced metabolism adaptation. Among the metabolic responses to oxidative stress, the expression of catalases, DnaJ- protein, and ATPase_ protein (Table 1), which were physiological responses induced by the metal, can be highlighted [26]. It is well known that under copper stress occur reduction in the expression of mitochondrial-related proteins [26]. However, increases in HSPs and catalase proteins were observed, conserving the cell homeostasis. This protein can be expressed or overexpressed under stress conditions, as shown in Table 1. Therefore, proteins identified in the presence of the metal, including binding proteins and cytochromes, led to a new structural composition in the fungal wall [23]. In this way, the expression of HSPs may be responsible for resistance to toxicity or absorption of the metal.
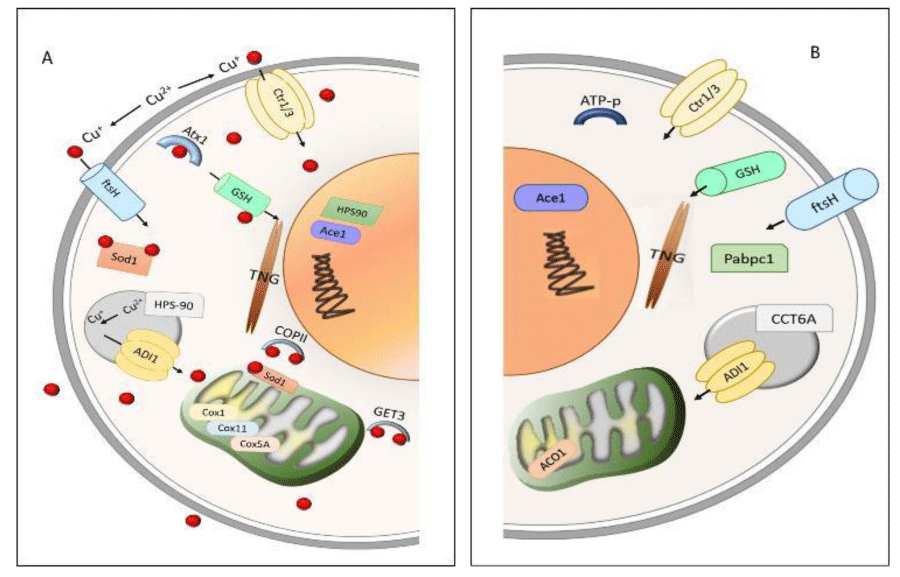
Small COPII coat GTPase is related to transmembrane copper transport and cellular copper tolerance (Figure 8). To avoid the oxidant metal effect during the cell depuration process, the copper-binding to SOD is transported until storing Get3, Cox, and HSP (Figure 8). In presence of metal, fungal metabolism showed ATPase/Get3 expression associated with cell uptake of copper ions. HSP protein was described as indicative of increased sensitivity to heat and oxidative stress [27]. Copper treatment increased the isoforms of HSP-7, HSP60, HSP70, and HSP78 proteins compared to control, indicating a metabolic response through HSPs expression, suggesting that copper stress modified the process of cellular modeling and adhesion in R. microspores. In another way, the presence of HSP90 only in the copper-treated group was suggested to maintain the protein expression stability. This adaptation is triggered by environmental stress to heat or cell wall damage, activating key transcription factors and intracellular signal transduction proteins [28].
Further, Cox protein as a final receptor of transporting metal from membrane to mitochondria was associated with SOD, suggesting that intracellular transport of copper ions was performed to complex Cox-SOD. Copper presence associated with the Cox1 subunit in mitochondria was measured by the copper joint action of the proteins Cox11 and Cox5A [29].
The overexpression of Get3, Cox, and HSP proteins was related to copper concentration in the cells. These proteins support the copper transport (absorption and desorption) between the cell and the environment. The desorption process observed in R. microsporus was also observed in A. niger IOC 4687 [18].
Therefore, the expression of new isoforms of HSPs in R. microsporus IOC 4686 suggests a cellular adaptation response to stress caused by exposure to copper. Changes in cell metabolism and metalloproteins expression must be by metal catalytic oxidation, which links divalent ions to a polypeptide chain and generates reactive oxygen species causing damage in adjacent amino acid chains [7,30,31].
Seven metalloproteins were identified in both, control and copper-treated, in the presence of copper, metalloproteins such as ATP-metallopeptidase Hfl, cytochrome P450, methionine aminopeptidase, and cell division protease fish were observed (Table 2, Figure 8). Changes in the metabolic pathways induced by the metals (stress) cause alteration in the cellular activity of the organisms, which results in disorders in cell homeostasis [30].
Considering genetic proximity from other species from Rhizopus genera (Figure 1) was observed that R. oryzea fungus in aqueous copper solution removed copper ions by viable and NaOH treated hyphae, being maximum copper loading capacity of the viable and pretreated biomass according to Langmuir isotherm was 19.4 and 43.7mgg-1, respectively [32].
In another way, the R. oryzea fungi consortium used for bioremediation of metals and dyes of textile industry effluents showed copper ions absorption of 25mgL-1 [33]. Further, R. stolonifer consortium in a medium contaminated by a mixture of Ni, Pb, and Cd showed high uptake efficiency of Pb (541.5mgkg-1) and Ni (501.05mgkg-1) [34]. Despite, copper recovery by R. microsporus from solution was not reported, this study showed that maximum copper recovery by IOC 4686 was higher than R. oryzea consortium [26]. Thus, intracellular and extracellular proteins conferring to fungus metal sequester and precipitation ability suggest that these fungi could be used for metal recovery from the contaminated environments [8,9].
The copper absorption process by Rhizopus arrhizus showed absorption of copper ions (640mgL-1), being weakly influenced by temperature ranging from 4 to 25ºC, and showed directly related metal absorption with biomass concentration [35]. Same fungi specie treated with Cd (II), Pb (II), and Cu (II) ions at different temperatures and pH, showed a maximum absorption concentration of 7.32mgg-1 Cu (II) in 72 h of cultivation at 30ºC and pH 4.5 [36] R. arrehizus also was studied in bioremediation processes in different copper concentrations, pH and temperature, showing 97.32% of maximum efficiency in culture containing 100mgL-1 of copper in pH 7.0 and 35°C [37]. Despite reports showing that the pH and temperature influenced metal recovery, this research does not fix temperature and pH, using similar conditions concerning the local sampling environment and attempting to lower bioremediation process costs.
Proteomic analyses of R. microsporus IOC 4686 incubated in a copper solution showed an increase in translocation membrane, oxidative molecular reaction, and metal-binding proteins compared to control. P450 was the main protein responsible for conferring resistance to the fungus under stress induced by copper. These proteins involved in the defense mechanism were found in different isoforms in copper-treated samples. Based on these results, R. microsporus IOC 4686 has the potential for metal bioremediation with a resistance mechanism capable of neutralizing oxidative stress induced by copper. The purpose of this study was to understand the mechanism of the fungus, R. microporus to absorb and accumulate metal from contaminated sites. This work was performed only with copper ions, but it could help to understand the physiological changes induced by other metals, such as Cr and Pb.
The Challenge of this work was to understand the changes in the metabolism of R. microsporus IOC 4686 induced by copper allowing environmental adaptation.
Author contribution
MD, ER, CN, and MM conceived and designed research. MD and LA conducted experiments. JL analyzed the data. MD, LA, and MM wrote the manuscript. All authors read and approved the manuscript.
Funding
This study was supported by a donation from the Brazilian Council of Science of Technological Development INCT. We also thank the FAPESP Proc. 2013/50218-2.
Data availability Sequence
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
- Ha W, Sevim-Nalkiran H, Zaman AM, Matsuda K, Khasraw M, Nowak AK, Chung L, Baxter RC, McDonald KL. Ibudilast sensitizes glioblastoma to temozolomide by targeting Macrophage Migration Inhibitory Factor (MIF). Sci Rep. 2019 Feb 27;9(1):2905. doi: 10.1038/s41598-019-39427-4. PMID: 30814573; PMCID: PMC6393433.
- Fomina M, Gadd GM. Biosorption: current perspectives on concept, definition and application. Bioresour Technol. 2014 May;160:3-14. doi: 10.1016/j.biortech.2013.12.102. Epub 2014 Jan 3. PMID: 24468322.
- DNPM. Sumário Mineral de 2015. Dep Nac Prod Min. 2020.
- Dias M, Gomes de Lacerda JTJ, Perdigão Cota de Almeida S, de Andrade LM, Oller do Nascimento CA, Rozas EE, Mendes MA. Response mechanism of mine-isolated fungus Aspergillus niger IOC 4687 to copper stress determined by proteomics. Metallomics. 2019 Sep 1;11(9):1558-1566. doi: 10.1039/c9mt00137a. Epub 2019 Sep 4. PMID: 31482901.
- Andrade LM, Tito CA, Mascarenhas C, Lima FA, Dias M, Andrade CJ, Mendes MA, Nascimento CAO. Chlorella vulgaris phycoremediation at low Cu+2 contents: Proteomic profiling of microalgal metabolism related to fatty acids and CO2 fixation. Chemosphere. 2021 Dec;284:131272. doi: 10.1016/j.chemosphere.2021.131272. Epub 2021 Jun 29. PMID: 34323785.
- Pele MA, Montero-Rodriguez D, Rubio-Ribeaux D, Souza AF, Luna MAC, et al. Development and improved selected markers to biosurfactant and bioemulsifier production by Rhizopus strains isolated from Caatinga soil. African J Biotechnol 2018; 17:150-157.
- Irazusta V, Estévez C, Amoroso MJ, de Figueroa LI. Proteomic study of the yeast Rhodotorula mucilaginosa RCL-11 under copper stress. Biometals. 2012 Jun;25(3):517-27. doi: 10.1007/s10534-012-9531-0. Epub 2012 Mar 6. PMID: 22391792.
- De Freitas AC, Escaramboni B, Carvalho AFA, De Lima VMG, De Oliva-Neto P. Production and application of amylases of Rhizopus oryzae and Rhizopus microsporus var. oligosporus from industrial waste in acquisition of glucose. Chem Pap. 2013; 68:442-450.
- Oladipo OG, Awotoye OO, Olayinka A, Bezuidenhout CC, Maboeta MS. Heavy metal tolerance traits of filamentous fungi isolated from gold and gemstone mining sites. Braz J Microbiol. 2018 Jan-Mar;49(1):29-37. doi: 10.1016/j.bjm.2017.06.003. Epub 2017 Aug 8. PMID: 28844883; PMCID: PMC5790576.
- The UniProt Consortium. UniProt: the universal protein knowledgebase. Nucleic Acids Res. 2017 Jan 4;45(D1):D158-D169. doi: 10.1093/nar/gkw1099. Epub 2016 Nov 29. Erratum in: Nucleic Acids Res. 2018 Mar 16;46(5):2699. PMID: 27899622; PMCID: PMC5210571.
- Alves LA, Souza RC, da Silva TM, Watanabe A, Dias M, Mendes MA, Ciamponi AL. Identification of microorganisms in biofluids of individuals with periodontitis and chronic kidney disease using matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Rapid Commun Mass Spectrom. 2016 May 30;30(10):1228-1232. doi: 10.1002/rcm.7555. PMID: 28328023.
- Küçükgöze G, Alkım C, Yılmaz Ü, Kısakesen Hİ, Gündüz S, Akman S, Çakar ZP. Evolutionary engineering and transcriptomic analysis of nickel-resistant Saccharomyces cerevisiae. FEMS Yeast Res. 2013 Dec;13(8):731-46. doi: 10.1111/1567-1364.12073. Epub 2013 Oct 3. PMID: 23992612.
- Dias M, Gomes de Lacerda JTJ, Perdigão Cota de Almeida S, de Andrade LM, Oller do Nascimento CA, Rozas EE, Mendes MA. Response mechanism of mine-isolated fungus Aspergillus niger IOC 4687 to copper stress determined by proteomics. Metallomics. 2019 Sep 1;11(9):1558-1566. doi: 10.1039/c9mt00137a. Epub 2019 Sep 4. PMID: 31482901.
- Olsson L, Nielsen J. On-line and in situ monitoring of biomass in submerged cultivations. Trends Biotechnol. 1997; 15:517-522.
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248-54. doi: 10.1006/abio.1976.9999. PMID: 942051.
- Lima KO, da Costa de Quadros C, Rocha MD, Jocelino Gomes de Lacerda JT, Juliano MA, et al. Bioactivity and bioaccessibility of protein hydrolyzates from industrial byproducts of Stripped weakfish (Cynoscion guatucupa). LWT 2019; 111.
- Zhang J, Xin L, Shan B, Chen W, Xie M, Yuen D, Zhang W, Zhang Z, Lajoie GA, Ma B. PEAKS DB: de novo sequencing assisted database search for sensitive and accurate peptide identification. Mol Cell Proteomics. 2012 Apr;11(4):M111.010587. doi: 10.1074/mcp.M111.010587. Epub 2011 Dec 20. PMID: 22186715; PMCID: PMC3322562..
- Perdigão Cota de Almeida S, Rozas EE, Oller do Nascimento CA, Dias M, Mendes MA. Metabolomic and secretomic approach to the resistance features of the fungus Aspergillus niger IOC 4687 to copper stress. Metallomics. 2021 Jan 21;13(1):mfaa010. doi: 10.1093/mtomcs/mfaa010. PMID: 33570139..
- Yuvaraj M, Udayakumar K, Jayanth V, Prakasa Rao A, Bharanidharan G, Koteeswaran D, Munusamy BD, Murali Krishna C, Ganesan S. Fluorescence spectroscopic characterization of salivary metabolites of oral cancer patients. J Photochem Photobiol B. 2014 Jan 5;130:153-60. doi: 10.1016/j.jphotobiol.2013.11.006. Epub 2013 Nov 25. PMID: 24333763.
- Mauch RM, Cunha Vde O, Dias AL. The copper interference with the melanogenesis OF Cryptococcus neoformans. Rev Inst Med Trop Sao Paulo. 2013 Mar-Apr;55(2):117-20. doi: 10.1590/s0036-46652013000200009. PMID: 23563765.
- Giansanti P, Tsiatsiani L, Low TY, Heck AJ. Six alternative proteases for mass spectrometry-based proteomics beyond trypsin. Nat Protoc. 2016 May;11(5):993-1006. doi: 10.1038/nprot.2016.057. Epub 2016 Apr 28. PMID: 27123950.
- Richter K, Haslbeck M, Buchner J. The heat shock response: life on the verge of death. Mol Cell. 2010 Oct 22;40(2):253-66. doi: 10.1016/j.molcel.2010.10.006. PMID: 20965420.
- Hagedoorn PL. Microbial Metalloproteomics. Proteomes. 2015 Dec 1;3(4):424-439. doi: 10.3390/proteomes3040424. PMID: 28248278; PMCID: PMC5217388.
- Li Q, Harvey LM, McNeil B. Oxidative stress in industrial fungi. Crit Rev Biotechnol. 2009;29(3):199-213. doi: 10.1080/07388550903004795. PMID: 19514862.
- de Santana FS, Gracioso LH, Karolski B, Dos Passos Galluzzi Baltazar M, Mendes MA, do Nascimento CAO, Perpetuo EA. Isolation of Bisphenol A-Tolerating/degrading Shewanella haliotis Strain MH137742 from an Estuarine Environment. Appl Biochem Biotechnol. 2019 Sep;189(1):103-115. doi: 10.1007/s12010-019-02989-0. Epub 2019 Mar 14. PMID: 30868384.
- Maio N, Kim KS, Singh A, Rouault TA. A Single Adaptable Cochaperone-Scaffold Complex Delivers Nascent Iron-Sulfur Clusters to Mammalian Respiratory Chain Complexes I-III. Cell Metab. 2017 Apr 4;25(4):945-953.e6. doi: 10.1016/j.cmet.2017.03.010. PMID: 28380382.
- Farkas Á, De Laurentiis EI, Schwappach B. The natural history of Get3-like chaperones. Traffic. 2019 May;20(5):311-324. doi: 10.1111/tra.12643. PMID: 30972921; PMCID: PMC6593721.
- Lamoth F, Juvvadi PR, Steinbach WJ. Heat Shock Protein 90 (Hsp90) in Fungal Growth and Pathogenesis. Curr Fungal Infect Rep. 2014; 8:296–301.
- Nevitt T, Ohrvik H, Thiele DJ. Charting the travels of copper in eukaryotes from yeast to mammals. Biochim Biophys Acta. 2012 Sep;1823(9):1580-93. doi: 10.1016/j.bbamcr.2012.02.011. Epub 2012 Feb 24. PMID: 22387373; PMCID: PMC3392525.
- Waldron KJ, Rutherford JC, Ford D, Robinson NJ. Metalloproteins and metal sensing. Nature. 2009 Aug 13;460(7257):823-30. doi: 10.1038/nature08300. PMID: 19675642.
- El Golli-Bennour E, Bacha H. Hsp70 expression as biomarkers of oxidative stress: mycotoxins' exploration. Toxicology. 2011 Sep 5;287(1-3):1-7. doi: 10.1016/j.tox.2011.06.002. Epub 2011 Jun 13. PMID: 21684316.
- Bhainsa KC, D'Souza SF. Removal of copper ions by the filamentous fungus, Rhizopus oryzae from aqueous solution. Bioresour Technol. 2008 Jun;99(9):3829-35. doi: 10.1016/j.biortech.2007.07.032. Epub 2007 Sep 4. PMID: 17804218.
- Mishra A, Malik A. Novel fungal consortium for bioremediation of metals and dyes from mixed waste stream. Bioresour Technol. 2014 Nov;171:217-26. doi: 10.1016/j.biortech.2014.08.047. Epub 2014 Aug 25. PMID: 25203229.
- Njoku KL, Akinyede OR, Obidi OF. Microbial Remediation of Heavy Metals Contaminated Media by Bacillus megaterium and Rhizopus stolonifer. Sci African. 2020; 10:e00545.
- de Rome L, Gadd GM. Copper adsorption by Rhizopus arrhizus, Cladosporium resinae and Penicillium italicum. Appl Microbiol Biotechnol. 26: 84-90.
- Aksu Z, Dönmez G. A comparative study on the biosorption characteristics of some yeasts for Remazol Blue reactive dye. Chemosphere. 2003 Mar;50(8):1075-83. doi: 10.1016/s0045-6535(02)00623-9. PMID: 12531715.
- Chauhan G, González-González RB, Iqbal HMN. Bioremediation and decontamination potentials of metallic nanoparticles loaded nanohybrid matrices - A review. Environ Res. 2022 Mar;204(Pt D):112407. doi: 10.1016/j.envres.2021.112407. Epub 2021 Nov 18. PMID: 34801543.
Article Alerts
Subscribe to our articles alerts and stay tuned.
 This work is licensed under a Creative Commons Attribution 4.0 International License.
This work is licensed under a Creative Commons Attribution 4.0 International License.


 Save to Mendeley
Save to Mendeley
