Open Journal of Analytical and Bioanalytical Chemistry
Triosephosphate isomerase from baker’s yeast – ribozyme versus protein
ON Solovjeva*
Cite this as
Solovjeva ON (2020) Triosephosphate isomerase from baker’s yeast – ribozyme versus protein. Open J Anal Bioanal Chem 4(1): 020-028. DOI: 10.17352/ojabc.000020It was previously shown that in baker’s yeast Saccharomyces cerevisiae, transketolase can exist not only free, but in complex with RNA. The complex does not possess transketolase activity [N.K. Tikhomirova, G.A. Kochetov, A new method of isolation and a new form of transketolase from baker’s yeast, Biokhimiia 56 (1991) 1123-1130].
We discovered that this RNA is a ribozyme which catalyzes the interconversion of glyceraldehyde 3-phosphate (G3P) and dihydroxyacetone 3-phosphate (DHAP), i.e. acting as triosephosphate isomerase (TPI). It also catalyzes an unusual reaction of ribose 5-phosphate (R5P) decomposition to G3P and erythrose. TPI-ribozyme was found in baker’s yeast not only in complex with transketolase, but also in free form. Transketolase-RNA complex was easily isolated on an immunoaffinity column with antibodies to transketolase. TPI-ribozyme consists of 87 nucleotides and has a molecular weight of 26.6 kDa. The optimum of pH-activity is 7.5 for DHAP, 6.7 for R5P and 9.0 for G3P. Km and Vmax are accordingly 0.29 mM and 2.6 U/mg for DHAP, 22 mM and 0.65 U/mg for R5P, 0.05 mM and 4.3 U/mg at pH 7.6 and 0.11 mM and 16 U/mg at pH 9.0 for G3P. These kinetic characteristics are the same for free RNA and in the complex with transketolase. Ki for RNA binding to transketolase was 1.0 μM. Accordingly, the TPI-ribozyme performs a dual function – it shows TPI activity and blocks the work of transketolase, thereby switching the metabolic process to glycolysis. The location of the TPI-ribozyme gene is determined. Blocking the activity of transketolase by ribozyme may be of practical importance in medicine, particularly, in cancer therapy.
Abbreviations
TK: Transketolase; ThDP: Thiamine Diphosphate; TPI: Triosephosphate Isomerase; GAPDH: Glyceraldehyde 3-Phosphate Dehydrogenase; GPDH: Glycerol 3-Phosphate Dehydrogenase; ACN: Acetonitrile; DHAP: Dihydroxyacetone 3-Phosphate; G3P: Glyceraldehyde 3-Phosphate; R5P: Ribose 5-Phosphate
Introduction
Ribozymes were first reported in 1981 [1-3]. They catalyze a variety of reactions, along with protein enzymes. The ribozymes involved in the synthesis and cleavage of RNA [4-7] and protein [8] are well studied. RNA ligase ribozyme (DSL) [9,10], Tetrahymena ribozyme [11,12], RNA polymerase ribozyme [13] and others are involved in the cleavage of RNA. In biosynthesis of amino acids is involved glmS ribozyme [14-16]. There are ribozymes that catalyze amino acids transfer reactions [17].
Ribozymes with peroxidase activity [18] and urea synthase activity [19] were detected. Self-cleaving ribozymes are also detected [20-23]. In addition, the ribosomes function as ribozymes [24-26], the RNase works in a protein complex with a ribozyme [27,28]. Among the ribozymes that convert carbohydrate substrates are the Diels-Alderase ribozyme, which is involved in the formation of C–C bonds [29,30], and the ribozyme that catalyzes the synthesis of dipeptides [31]. About 100 reviews have been written about these ribozymes. The review [32], contains a table of RNA classes, their sizes and functions. The review [33], provides a detailed description of the chemical properties of ribozymes, the role of metals and the characteristics of individual ribozymes, as well as the methods used in ribozyme studies. Self-cleaving ribozymes are described in review [23], riboswitches in reviews [34,35]. The latest review of RNase was published in 2009 [28].
Since 2003, 5 articles have been published on ribozymes that are not involved in the biosynthesis of RNA or protein. They are involved in the reactions of alcoholic fermentation - pyruvate decarboxylase ribozyme [36] and alcohol dehydrogenase ribozyme [37,38] and in glycolysis - aldolase ribozyme [39,40]. Interestingly, free thiamine is used in the pyruvate decarboxylase ribozyme, while thiamine diphosphate is used in the pyruvate decarboxylase protein [36]. Alcohol dehydrogenase ribozyme oxidizes alcohol only in the presence of NAD+ and Zn2+ [37]. Like alcohol dehydrogenase, a protein, it is able to catalyze a reverse reaction in the presence of NADH and Zn2+ [38]. Aldolases (protein enzymes) utilize either an enamine mechanism or a Zn2+ cofactor. Aldolase ribozyme is a Zn2+- dependent. Ribozyme, unlike the protein enzyme, can also catalyze a transfer of a biotinylated benzaldehyde derivative to the aldol donor substrate [39]. All were obtained by in vitro selection using an RNA library.
In this paper, we first show the existence of another glycolytic ribozyme, Triosephosphate Isomerase (TPI)-ribozyme, which was found in baker’s yeast, both free and associated with Transketolase (TK), performing, respectively, two functions - the work of the glycolytic enzyme and blocking the enzyme of the pentose phosphate pathway and accordingly the switching of metabolism to glycolysis. The activity of TPI-ribozyme is the same as a part of the complex and in free form. It was previously shown that TK can be isolated from yeast in the form of a complex with RNA [41,42]. G3P, which is formed from DHAP upon catalysis by TPI (enzyme and ribozyme), is one of the substrates of transketolase. Nevertheless, it is not clear why the shutdown of TK is carried out precisely by TPI-ribozyme.
We suggest that in perspective for most enzymes, ribozymes with a similar function will be found. It is possible that other ribozymes will also be associated with proteins and so will be easy to isolate. The presence of a complex with nucleic acids is known for many enzymes, including those not involved directly in the biosynthesis of nucleic acids and proteins [43-45]. For glyceraldehyde 3-phosphate dehydrogenase (GAPDH), the interaction with the hepatitis delta virus RNA was shown [46,47].
Materials and methods
Materials
Thiamine Diphosphate (ThDP), dithiothreitol, glycylglycine, MES were from Serva, NAD+, NADH, CaCl2, GAPDH, glycerol 3-phosphate dehydrogenase (GPDH), RNAse A, Dihydroxyacetone Phosphate (DHAP), Ribose 5-Phosphate (R5P), erythrose, CHES, acetonitrile (ACN), perchloric acid, ammonium sulfate, protamine sulfate from Sigma, HEPES from Fluka, human albumin, pepsin and lysozyme from Reanal, other commercially available chemicals were of the highest grade.
Transketolase purification
TK (free enzyme and in complex with RNA) was isolated from baker’s yeast Saccharomyces cerevisiae on a column with immobilized antibodies to TK isolated from the serum of an immunized rabbit according to the method described previously [42]. Extraction was carried out by cytolysis of dry yeast with 0.5 M NH4OH. During the elution of TK from the column, the pH was maintained at 7.6 by adding 4M HCl to the eluate. The eluted protein was concentrated in Amicon® 50,000 MWCO Ultra Centrifugal Filters, then aliquoted and stored frozen. Before use, the TK solution was passed through a Sephadex G-50 column equilibrated with 50 mM glycylglycine, pH 7.6.
Measuring protein and RNA concentrations
The concentration of free TK was determined spectrophotometrically using the absorption coefficient A 1% 1-cm of 14.5 at 280 nm [48]. The concentration of TK in TK-RNA complex was determined according to the Bradford method [49], using the free TK for calibration. The amount of nucleotides was calculated by [50], in which the optical density at 260 nm equals 1.0 for the 40 µg/mL solution of RNA, for a 1-cm pathlength. The molar concentration of nucleotides was calculated by taking an average nucleotide mass of 345 Da.
Purification of the TK-RNA complex
The TK isolated on the immune column (mixture of free TK and TK-RNA complex) was passed through a Sephadex G-50 column in 10 mM potassium phosphate buffer, pH 7.6. Then, 2 mL of TK + TK-RNA was passed through the IRA-400 column (2.4 mL), previously washed with 100 mL of 0.1 M KOH and with water until neutral pH and eluted with the same buffer. Free enzyme was absorbed onto the column.
Obtaining free RNA from the TK-RNA complex
ACN in a 2:1 ratio was added to the TK-RNA complex, stirred, and frozen for 1-2 h at -12oC. The upper layer of ACN, which did not contain RNA, was removed, in the lower layer of water, the denatured protein was removed by centrifugation. The RNA obtained in this way was precipitated with 96% ethanol (3:1) and dissolved in water. Determination of protein by Bradford showed its absence.
Obtaining free TK from the TK-RNA complex
ApoTK (TK without ThDP and RNA) was obtained by keeping TK (2mg/mL) in 1.6M ammonium sulfate at pH 7.6 for 24h at 4oC. Then the enzyme was precipitated with 4 M ammonium sulfate, the precipitate was dissolved in 50 mM glycylglycine or potassium phosphate buffer, pH 7.6, and passed through a Sephadex G-50 column equilibrated with the same buffer.
Determination of RNA molecular mass
Initially, the molecular mass of RNA was determined by gel chromatography on a Sephadex G-100 column in 6 M urea. A column of 15 x 1 mL was applied to 1 mL of RNA. Human albumin 66.44 kDa, pepsin 34.5 kDa, and lysozyme 14.5 kDa were taken as markers. All compounds were kept for 2 h in urea before being applied to the column.
Then, the molecular mass of RNA was determined using MALDI-TOF MS analysis, which was performed on an UltrafleXtreme MALDI-TOF/TOF mass spectrometer (Bruker Daltonics, Germany) equipped with an Nd laser by detection of MH+ molecular ions.
Determination of thiamine diphosphate concentration
The concentration of ThDP was determined spectrophotometrically by measuring optical density at 272.5 nm (molar extinction coefficient 7500 M−1 ·cm−1) [51] and in TK-RNA - enzymatically, after boiling or incubation with RNase A [52].
Measuring of transketolase activity
The catalytic activity of TK was measured at room temperature using spectrophotometer Ultrospec 110 pro by the rate of NAD+ reduction in a coupled system with GAPDH [53]. The reaction mixture in final volume 1 mL contained 50 mM glycylglycine pH 7.6, 2.5mM CaCl2, 1 mM sodium arsenate, 3 mM dithiothreitol, 0.1 mM ThDP, 1.6 mM NAD+, 3 units of GAPDH and 3.5 mg/mL potassium salt of the phosphopentose mixture that was used as substrate. The reaction was initiated by the addition of TK. Phosphopentose mixture was obtained from R5P [54].
Determination of the affinity of RNA to TK
50 μg/mL of TK were incubated without RNA and with fixed RNA concentrations of 0.55, 1.0, and 2.65 μM and varying ThDP concentrations of 0.6-3 μM for 40 min without adding cations to the medium. Under these conditions, Ca2+ is located in only one active site of the dimeric TK molecule. Therefore, ThDP can be bound with only one of the two active sites [55,56]. The results were processed in reverse coordinates. Ki was calculated using the formula Km [I] / (Km app - Km).
Also, 0.5 mg/mL TK (3.4 μM) was incubated for 40 min with 6 μM ThDP without the addition of cations (condition of ThDP binding in one of two active sites). Measuring the activity without the addition of cofactors, we were convinced that it is 50% of the maximum, i.e. ThDP was bound only in the first active site. 6 μM RNA was then added and incubated for 20 min and the activity of TK was measured in the presence of 0.1 mM Ca2+ and 0.1 mM ThDP.
Measuring of RNA and TPI (protein) activity
The catalytic activity of free RNA and in the complex with TK, as well as the activity of the TPI - protein enzyme was measured by the method [57].
When the activity was measured using DHAP, the reaction mixture in the final volume of 1 mL contained 50 mM glycylglycine, 1.6 mM NAD+, 1 mM DHAP, 3 units GAPDH, 6 mM sodium arsenate, 3 mM dithiothreitol, 0.5-2.5 µg/mL RNA, pH 7.6.
When the activity was measured using glyceraldehyde 3-phosphate (G3P), the reaction mixture in the final volume of 1 mL contained 50 mM glycylglycine, 0.3 mM NADH, 1 mM G3P, 3 units of GPDH, 0.5-2.5 µg/mL RNA, pH 7.6.
When the activity was measured using DHAP or G3P, their freshly prepared solutions were used. The reaction was started by adding TK-RNA or RNA, after waiting for a zero background.
When the activity was measured using R5P, the reaction mixture in the final volume of 1 mL contained 50 mM glycylglycine, 0.3 mM NADH, 3 mM R5P, 3 units of GPDH, 0.5-2.5 µg/mL RNA, pH 7.6.
Km for substrates was measured in the concentration range of DHAP 0.03-2.2 mM, G3P 0.015-0.3 mM, R5P 2.5-120 mM. Experimental data were analyzed using the Michaelis equation
v = V · [S] / ([S] + Km)
When measuring the pH optimum, 50 mM buffers were used - MES at pH range 5.0-7.0, HEPES at pH range 7.0-8.5 and CHES at pH range 8.5-10.5.
Erythrose assay
After the total conversion of R5P by RNA in the presence of GPDH and NADH, the reaction mixture was treated with perchloric acid. The precipitated protein was removed by centrifugation, and the supernatant was assayed for the amount of erythrose in the cysteine - H2SO4 reaction [58].
Isolation of total RNA from yeast extract
Extraction was carried out from dry yeast with 0.5 M ammonia, as in the isolation of TK. Half of the extract was passed through an anti-TK immunoaffinity column to bind and thus remove the TK and the TK-RNA complex. ACN 3:1 was added to both extracts to remove proteins.
Partial purification of TPI - protein enzyme
After removing the nucleic acids from the yeast extract by treatment with protamine sulfate and removing the TK-RNA by passing through an immunoaffinity column with antibodies to TK, dry ammonium sulfate was added to 70% saturation. The precipitate was collected, dissolved in the same volume, and the protein fraction was collected with saturation of 60-70% ammonium sulfate. The absence of GAPDH activity was verified [59].
RNA sequence
RNA was extracted from RNA-protein complex with PureLink RNA Micro Kit (Invitrogen) according to manufacture instruction. NGS Illumina library was made from RNA with NEBNextNEBNext® Ultra™II RNA Library Prep Kit for Illumina® (NEB) according to manual. Sequencing was performed on HiSeq1500 with 50 bp read length. 13 millions of reads were generated for each sample and mapped to reference genome Saccharomyses cerevisiae. Read counts to every transcripts was calculated.
Results and discussion
Isolation of TK-RNA on a column with IRA-400
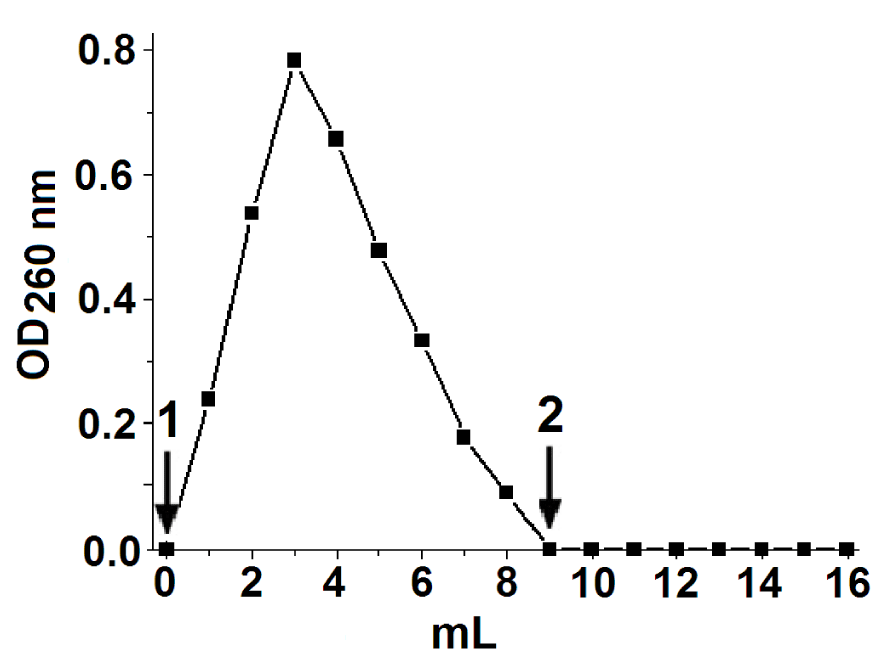
To obtain the TK-RNA complex without free TK, the TK isolated on the immunoaffinity column was passed through the column IRA-400. TK-RNA was eluted with 10 mM potassium phosphate buffer at pH 7.6. Free TK remained bound to the ion exchanger even when passing 500 mM buffer through the column (Figure 1). The TK-RNA complex did not possess transketolase activity, which was also shown earlier [41,42]. ThDP in the complex was absent (It was measured enzymatically). RNA can be separated from TK-RNA by ammonium sulfate and therefore is associated non-covalently.
Absorption spectra of TK-RNA and RNA
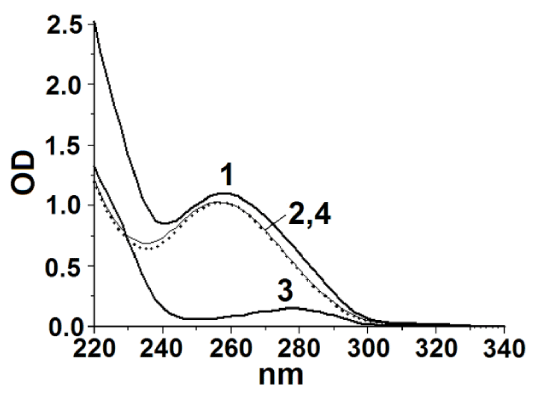
Figure 2 shows the absorption spectra of TK-RNA (curve 1), RNA obtained from TK-RNA by protein precipitation using ACN (curve 2), free TK (curve 3). In all samples, the concentration of TK was 0.1 mg/mL. The protein concentration in TK-RNA complex was determined according to Bradford [49]. The correctness of the determination was confirmed by the fact that in the difference spectrum (TK-RNA - TK) (curve 4) a spectrum was obtained that almost completely coincided with the spectrum of RNA (curve 2).
Measuring the activity of TK-RNA in the usual for TK transferase reaction in the presence of cofactors showed its complete absence. This means that RNA was associated with both active sites of TK, that is, two RNA molecules are linked to one dimeric TK molecule. Comparing the concentration of TK and nucleotides in the TK-RNA complex, we calculated that one RNA molecule contains 87 nucleotides, i.e. the mass of RNA is about 30 kDa. The molar extinction coefficients calculated from the absorption spectra at different wavelengths are shown in Table 1. A280 for TK bound to RNA is 6.03, while for free TK this value is 1.45 [48].
Determination of the RNA molecular mass
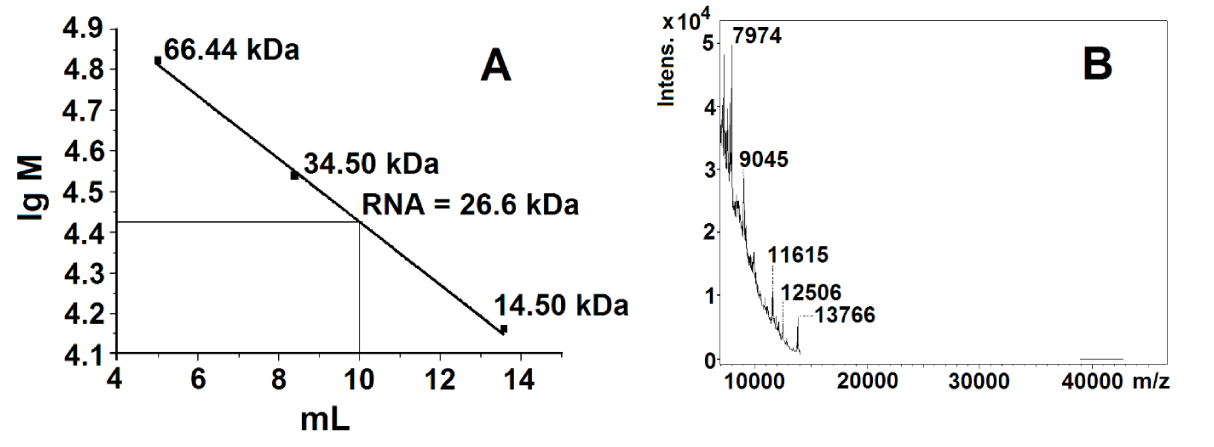
The molecular mass of RNA determined by gel chromatography on Sephadex G-100 under denaturing conditions was 26.6 kDa (Figure 3A). Then, the molecular mass of RNA was determined using MALDI-TOF MS analysis. The maximum mass value of 13.766±2 kDa was determined (Figure 3B). So, it can be assumed that the RNA is double-stranded. These data require further research.
Determination of RNA affinity for TK
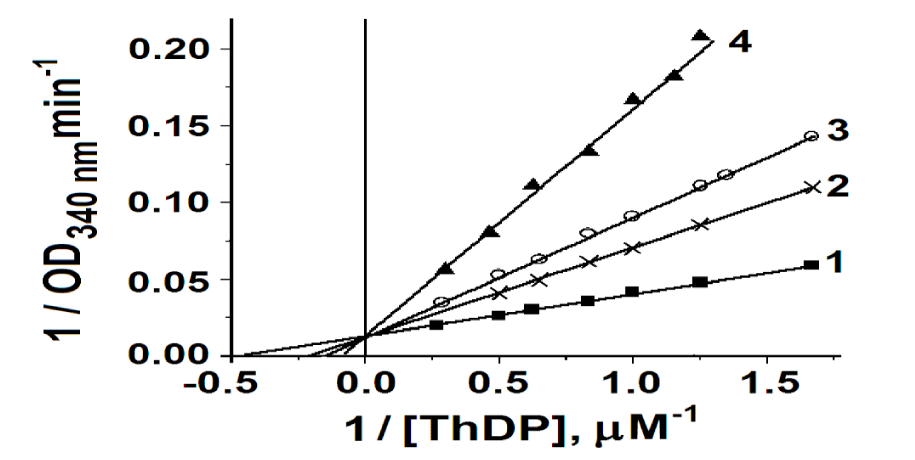
ApoTK was incubated with varying concentrations of ThDP in the presence of fixed concentrations of RNA and in its absence to determine the type of inhibition and Ki upon binding of RNA to TK. Binding was performed without the addition of cations in the incubation samples and then the TK activity was measured without the addition of cofactors. As it was shown earlier [51,52] in the absence of added cations from the outside, Ca2+ is bound in one of two active sites of TK and, accordingly, ThDP can only be linked in the first active site. In Figure 4 it is shown that in this case competitive binding of RNA and ThDP occurs. This means that RNA binds to or near the active site of the TK. Under these conditions, binding of RNA can only be seen in the first active site, according to the decrease in TK activity. Ki for RNA binding was 1.0 ± 0.2 μM, Kd for ThDP - 2.4 ± 0.2 μM.
In the second active site, which does not contain a cation, RNA binding also occurs. This was shown as follows. In the absence of cations in the medium, a small excess of ThDP was added to apoTK, with the result that ThDP was bound only in the first active site, and, accordingly, the enzyme activity was 50% of the activity measured in the presence of Ca2+ and ThDP. Then an excess of RNA was added to this enzyme. Subsequent measurement of TK activity with the addition of Ca2+ and ThDP showed that it was still 50% of the maximum. This means that RNA was bound to half of the active sites and that no cation is needed for its binding.
If the binding of RNA was performed in the presence of Ca2+ in the medium, it did not occur. Thus, RNA does not bind to TK in the presence of Ca2+ in the medium but it binds in the presence of Ca2+ in one active site. Besides, RNA does not displaced by Ca2+ and ThDP from the TK-RNA complex (data not shown).
Thus, RNA binds to TK in the region of its active sites competitively with respect to ThDP. Cations are not needed for RNA binding. After RNA treatment with RNase A, ThDP was not detected in it (data not shown). Previously, it was shown that the acceptor substrates bind to holoTK through the residue Asp477, which forms a hydrogen bond with the C2-hydroxyl group of the acceptor substrate [60]. Therefore, it can be assumed that RNA also binds directly to TK (perhaps also with the residue Asp477) through its R5P.
Reactions catalyzed by RNA. RNA is TPI-ribozyme
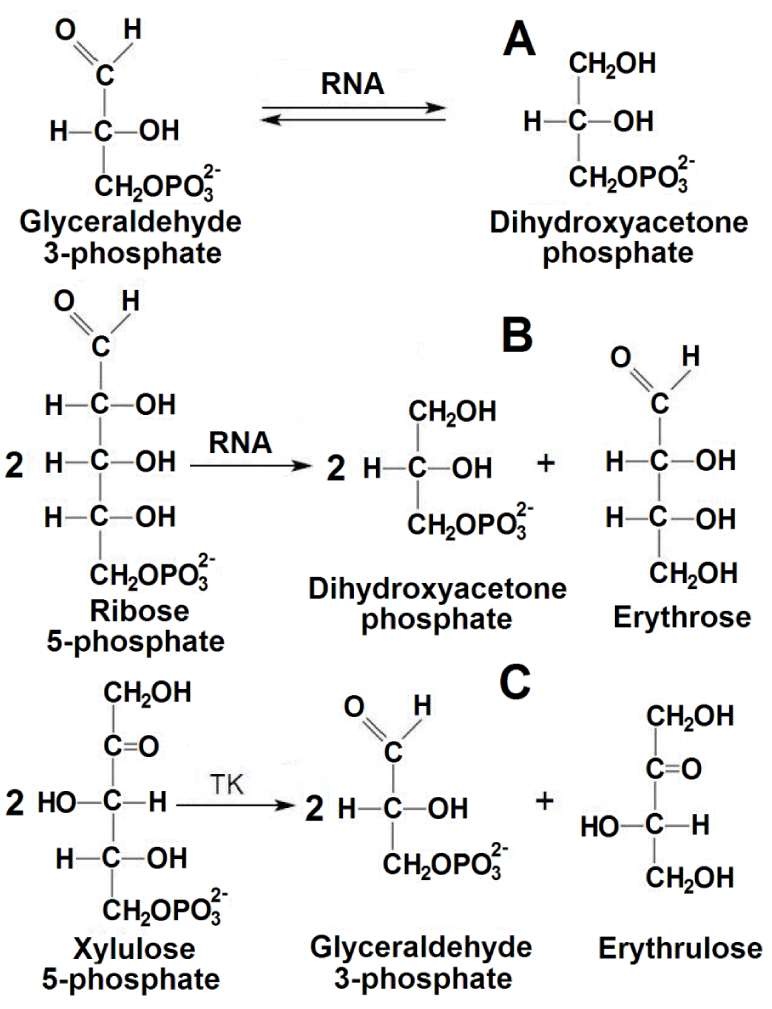
RNA associated with TK catalyzes the interconversion reaction of G3P and DHAP, like the protein enzyme TPI, as well as the unusual reaction of converting R5P to DHAP and erythrose. So, it is a ribozyme.
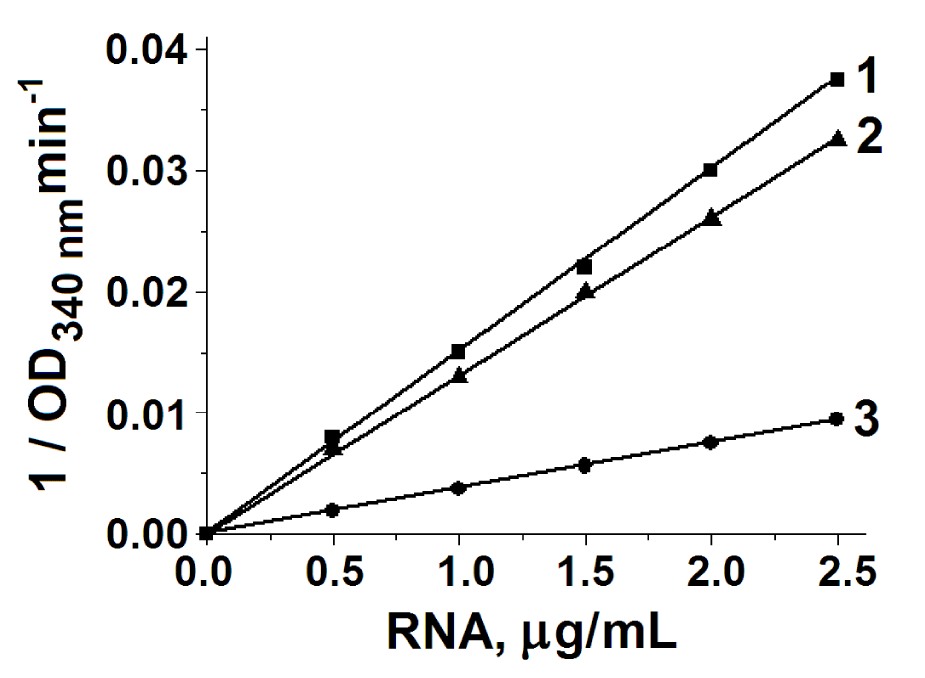
In Figure 5 it is shown the dependence of the rate of these reactions on the concentration of RNA. The reaction rate and kinetic characteristics were the same for free RNA and RNA in a complex with TK. The values of Km and Vmax for the reactions with these three substrates are shown in Table 2.
In the reaction of RNA with a five-carbon R5P, a three-carbon G3P is formed. Consequently, it should be expected that glycolaldehyde will be the second reaction product. However, the second product of the reaction was erythrose, which may be formed as a result of the condensation of two glycolaldehyde molecules. Assuming that this is the case, two molecules of RNA-converted R5P would yield one molecule of erythrose, as was confirmed experimentally. Table 3 shows that the amount of erythrose formed corresponds to the calculated data.
In Scheme 1 are shown the schemes of reactions with RNA and of the one-substrate reaction with TK. It is interesting that in one-substrate reaction of free TK with xylulose 5-phosphate, G3P is released in the medium and erythrulose is formed from the two molecules of glycolaldehyde [61], (Scheme 1C). For RNA, when R5P is used, two molecules of DHAP are sequentially released in the medium, and erythrose is formed from the two molecules of glycolaldehyde (Scheme 1B). So, when the ketose xylulose 5-phosphate is converted, the aldose G3P and the ketose erythrulose are formed. When the aldose R5P is converted, the ketose DHAP and the aldose erythrose are formed. It is interesting that two molecules of glycolaldehyde can form both ketose and aldose. Since the formation of erythrose on RNA occurs stoichiometrically, the first molecule of glycolaldehyde formed is not released into the medium, but remains bound to RNA, similarly to the same process in the case of a one-substrate reaction with TK.
The optimum pH in the reactions of RNA with G3P, DHAP and R5P
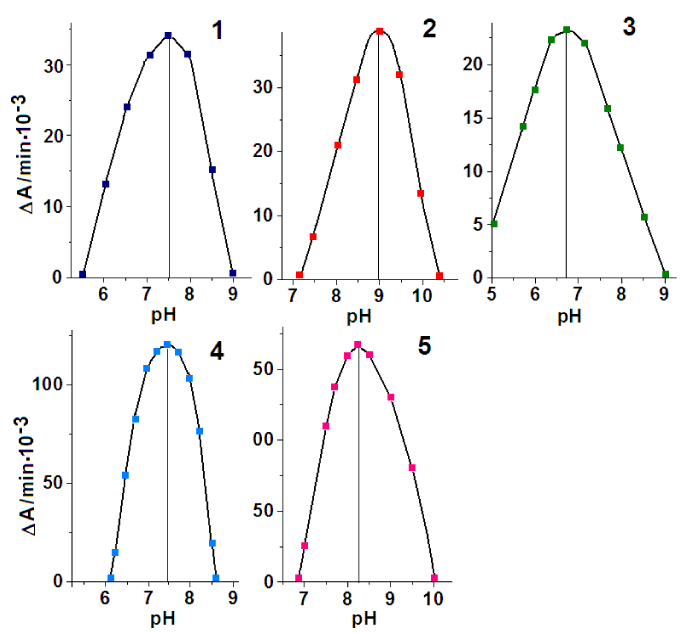
In Figure 6 are shown the results of the measuring the optimum pH activity in the reactions of RNA with substrates. The optimum pH in the reaction of RNA with DHAP is 7.5 (curve 1), with G3P 9.0 (curve 2), with R5P 6.7 (curve 3).
Comparison of the properties of TPI-ribozyme – free and in complex with TK and TPI-protein in yeast extract
Total RNA was isolated from yeast extract. A portion of the RNA was passed through an immunoaffinity column to remove the TK-RNA complex. Before and after removal of TK-RNA from the extract, there was RNA activity with G3P, DHAP and R5P. Consequently, TPI-ribozyme is in the extract not only in the form of a complex with TK, but also in a free form.
An ammonium sulfate fraction containing TPI (protein) was also obtained from the yeast extract and the kinetic constants were measured. The ratio of direct and reverse Km reactions is different for RNA and protein: for RNA, the affinity for G3P is 6 times higher than for DHAP, and for protein, the affinity for DHAP is 3 times higher than for G3P. At the same time, the affinity for G3P is 40 times higher in RNA than in protein (Table 2). Only RNA has the ability to catalyze the reaction with R5P. Vmax of the salt fraction containing TPI was 20 U/mg. Previously, it was shown that the specific activity of TPI from brever’s yeast was 41 U/mg in crude homogenate and 10,000 U/mg in the final purification stage [62]. Km for TPI (enzyme) of two types of yeast is almost the same for both substrates (Table 2).
The optimum pH of the reactions for RNA and TPI (enzyme) is approximately the same - 7.5 in the reaction with DHAP (Figure 7, curves 1 and 4) and 9.0 and 8.3 in the reaction with G3P for RNA and protein, respectively (Figure 7, curves 2 and 5).
RNA sequence
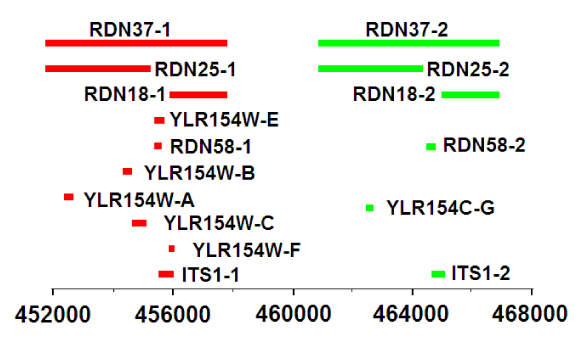
An RNA sequence was performed. The results are presented in Table 4 and Scheme 2. The largest and with the same number of readings falls on the genes of the ribosomal RNA RDN37-1 and RDN37-2, located in the 12th chromosome and each 5947 nucleotides in length. Several genes that are located within the genes RDN37-1 and RDN37-2 are also sequenced. These are genes RDN25-1 and RDN25-2 of 3396 nucleotides each and RDN18-1 and RDN18-2 of 1600 nucleotides each, as well as several short genes. It can be assumed that TPI-ribozyme is formed by splicing of some exons and is represented by two forms - TPI-ribozyme1 and TPI-ribozyme2, each about 87 nucleotides in length.
Conclusion
Previously was detected inactive TK from baker’s yeast [41,42]. TK activity was absent as a result of it’s binding to RNA and was restored when the TK-RNA complex dissociated. So, RNA binding is non-covalent. Due to the great interest in ribozymes, we tried to determine whether RNA associated with TK possesses enzymatic activity. As it turned out, this RNA is a TPI ribozyme and, in addition, unlike protein TPI, slowly cleaves R5P into DHAP and erythrose.
Currently, there are a lot of ribozymes with diverse functions. However, only ribozymes with peroxidase activity [18], urea synthase activity [19], pyruvate decarboxylase activity [36], alcohol dehydrogenase activity [37,38] and aldolase activity [39,40] which exhibit activity similar to enzymes, were found. There is no doubt that such ribozymes are actually much more.
In the case of TK, TPI-ribozyme not only duplicates the work of the protein enzyme, but also blocks the work of TK, thus switching yeast metabolism from the pentose phosphate pathway to glycolysis. The lack of transketolase activity was seasonal. Maximum of complex TK-RNA was in December. The reason for this switch remains to be found. But this fact indicates that turning off the functioning of TK by forming its complex with RNA is a real mechanism for switching the pentose phosphate pathway and glycolysis in baker’s yeast.
We were unable to determine the nucleotide sequence of the RNA ribozyme. According to preliminary data, this RNA is formed from the loci of two genes - RDN37-1 and RDN37-2 which have the same number of nucleotides (Scheme 2 and Table 4). The fact that the mass of RNA determined on the mass spectrometer is 2 times less than that determined with using gel chromatography (13.283 and 26.6 kDa consequently), indicates the possibility that our RNA is double-stranded. Perhaps these are two ribozymes with the same function.
It is known that to suppress the growth of cancer cells, inhibition of TK is performed [63,64]. Blocking of TK activity could be carried out by forming its complex with RNA.
I thank M.V. Serebryakova for MALDI-TOF MS analysis of RNA.
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
- Cech TR, Zaug AJ, Grabowski PI (1981) in vitro splicing of the ribosomal RNA precursor of Tetrahymena: involvement of a guanosine nucleotide in the excision of the intervening sequence. Cell 27: 487-496. Link: https://bit.ly/31WhFyJ
- Kruger K, Grabowski PJ, Zaug AJ, Sands J, Gottschling DE, et al. (1982) Self-splicing RNA: autoexcision and autocyclization of the ribosomal RNA intervening sequence of Tetrahymena. Cell 31: 147-157. Link: https://bit.ly/2CyEekl
- Guerrier-Takada C, Gardiner K, Marsh T, Pace N, Altman S (1983) The RNA moiety of ribonuclease P is the catalytic subunit of the enzyme. Cell 35: 849-857. Link: https://bit.ly/3iQwBW2
- Lilley DM (2011) Mechanisms of RNA catalysis. Phil Trans R Soc B 366: 2910-29107. Link: https://bit.ly/31W5PEQ
- Lilley DM (2011) Catalysis by the nucleolytic ribozymes. Biochem Soc Trans 39: 641-646. Link: https://bit.ly/2Q5pWdT
- Wilson TJ, Lilley DM (2015) RNA catalysis – is that it? RNA 21: 534-537. Link: https://bit.ly/3h4Llju
- Müller S, Appel B, Balke D, Hieronymus R, Nübel C (2016) Thirty-five years of research into ribozymes and nucleic acid catalysis: where do we stand today? F1000Res 5: F1000 Faculty Rev-1511. Link: https://bit.ly/345T2lK
- Suga H, Cowan JA, Szostak JW (1998) Unusual metal ion catalysis in an acyl-transferase ribozyme. Biochemistry 37: 10118-10125. Link: https://bit.ly/2Y7sBZe
- Horie S, Ikawa Y, Inoue T (2006) Structural and biochemical characterization of DSL ribozyme. Biochem Biophys Res Commun 339: 115-121. Link: https://bit.ly/3iTdsTn
- Glasner ME, Bergman NH, Bartel DP (2002) Metal ion requirements for structure and catalysis of an RNA ligase ribozyme. Biochemistry 41: 8103-8112. Link: https://bit.ly/345IVNL
- Herschlag D, Eckstein F, Cech TR (1993) The importance of being ribose at the cleavage site in the Tetrahymena ribozyme reaction? Biochemistry 32: 8312-8321. Link: https://bit.ly/3kL8TvU
- Guo F, Gooding AR, Cech TR (2004) Structure of the Tetrahymena ribozyme: base triple sandwich and metal ion at the active site. Mol Cell 16: 351-362. Link: https://bit.ly/2Q1kid4
- Zaher HS, Unrau PJ (2007) Selection of an improved RNA polymerase ribozyme with superior extension and fidelity. RNA 13: 1017-1026. Link: https://bit.ly/31Xx2XI
- McCarthy TJ, Plog MA, Floy SA, Jansen JA, Juliane K, et al. (2005) Ligand requirements for glmS ribozyme self-cleavage. Chem Biol 12: 1221-1226. Link: https://bit.ly/31Wh2Fn
- Brooks KM, Hampel KJ (2011) Rapid steps in the glmS ribozyme catalytic pathway: cation and ligand requirements. Biochemistry 50: 2424-2433. Link: https://bit.ly/2Y8eK4Z
- Bingaman JL, Zhang S, Stevens DR, Yennawar NH, Hammes-Schiffer S, et al. (2017) The GlcN6P cofactor plays multiple catalytic roles in the glmS ribozyme nature. Chem Biol 13: 439-445. Link: https://bit.ly/2EcRgEl
- Lohse PA, Szostak JW (1996) Ribozyme-catalysed amino-acid transfer reactions. Nature 381: 442-444. Link: https://bit.ly/3h8FpG8
- Travascio P, Bennet AJ, Wang DY, Sen D (1999) A ribozyme and a catalytic DNA with peroxidase activity: active sites versus cofactor-binding sites. Chem Biol 6: 779-787. Link: https://bit.ly/2E988w0
- Nieuwlandt D, West M, Cheng X, Kirshenheuter G, Eaton BE (2003) The first example of an RNA urea synthase: selection through the enzyme active site of human neutrophile elastase. Chembiochem 4: 651-654. Link: https://bit.ly/3h3uRIv
- Tang J, Breaker RR (2000) Structural diversity of self-cleaving ribozymes. PNAS 97: 5784-5789. Link:
- Doherty EA, Doudna JA (2001) Ribozyme structures and mechanisms. Annu Rev Biophys Biomol Struct 30: 457-475. Link:
- Cochrane JC, Strobel SA (2008) Catalytic strategies of self-cleaving ribozymes. Acc Chem Res 41: 1027-1035. Link: https://bit.ly/3219HUS
- Lee KY, Lee BJ (2017) Structural and biochemical properties of novel self-cleaving ribozymes. Molecules 22: E678. Link: https://bit.ly/3434zSZ
- DeRose VJ (2002) Two decades of RNA catalysis. Chem Biol 9: 961-969. Link: https://bit.ly/3awbAwB
- Kingery DA, Pfund E, Voorhees RM, Okuda K, Wohlgemuth I, et al. (2008) An uncharged amine in the transition state of the ribosomal peptidyl transfer reaction. Chem Biol 15: 493-500. Link: https://bit.ly/3iOAzhS
- Wohlgemuth I, Brenner S, Beringer M, Rodnina MV (2008) Modulation of the rate of peptidyl transfer on the ribosome by the nature of substrates. J Biol Chem 283: 32229–32235. Link: https://bit.ly/3iOOqok
- Kikovska E, Svard SG, Kirsebom LA (2007) Eukaryotic RNase P RNA mediates cleavage in the absence of protein. Proc Natl Academ Sci USA 104: 2062-2067. Link: https://bit.ly/3gYyijv
- Marvin MC, Engelke DR (2009) Broadening the mission of an RNA enzyme. J Cell Biochem 108: 1244-1251. Link: https://bit.ly/31WU94B
- Tarasow TM, Tarasow SL, Eaton BE (1997) RNA-catalysed carbon-carbon bond formation. Nature 389: 54-57. Link: https://bit.ly/2PV8Q2y
- Stuhlmann F, Jäschke A (2002) Characterization of an RNA active site: Interactions between a Diels-Alderase ribozyme and its substrates and products. J Am Chem Soc 124: 3238-3244. Link: https://bit.ly/348auGi
- Sun L, Cui Z, Gottlieb RL, Zhang B (2002) A selected ribozyme catalyzing diverse dipeptide synthesis. Chem Biol 9: 619-628. Link: https://bit.ly/2DQ37st
- Cech TR, Steitz JA (2014) The noncoding RNA revolution-trashing old rules to forge new ones. Cell 157: 77-94. Link: https://bit.ly/312X4JR
- Ward WL, Plakos K, DeRose VJ (2014) Nucleic acid catalysis: metals, nucleobases, and other cofactors. Chem Rev 114: 4318–4342. Link: https://bit.ly/346K0Fe
- Ramesh A, Winkler WC (2014) Metabolite-binding ribozymes. Biochim. Biophys Acta 1839: 989-994. Link: https://bit.ly/2Y66p1s
- Roßmanith J, Narberhaus F (2017) Modular arrangement of regulatory RNA elements. RNA Biol 14: 287-292. Link: https://bit.ly/3iK1awr
- Cernak P, Sen D (2013) A thiamin-utilizing ribozyme decarboxylates a pyruvate-like substrate. Nat Chem 5: 971-977. Link: https://bit.ly/344g6Bq
- Tsukiji S, Pattnaik SB, Suga H (2003) An alcohol dehydrogenase ribozyme. Nat Struct Biol 10: 713-717. Link: https://bit.ly/311P4bM
- Tsukiji S, Pattnaik SB, Suga H (2004) Reduction of an aldehyde by a NADH/Zn2+-dependent redox active ribozyme. J Am Chem Soc 126: 5044-5055. Link: https://bit.ly/2CDC572
- Fusz S, Eisenführ A, Srivatsan SG, Heckel A, Famulok M (2005) A ribozyme for the aldol reaction. Chem Biol 12: 941-950. Link: https://bit.ly/312t6FJ
- Fusz S, Srivatsan SG, Ackermann D, Famulok M (2008) Photocleavable initiator nucleotide substrates for an aldolase ribozymes. J Org Chem 73: 5069-5077. Link: https://bit.ly/3kQYj6K
- Tikhomirova NK, Merchan AY, Kochetov GA (1990) A new form of baker's yeast transketolase. An enzyme-RNA complex. FEBS Lett 274: 27-29. Link: https://bit.ly/2FobyLM
- Solovjeva ON (2002) Isolation and properties of noncovalent complex of transketolase with RNA. Biochemistry (Moscow) 67: 667-671. Link: https://bit.ly/2Cz0P01
- Routh SB, Sankaranarayanan R (2018) Enzyme action at RNA-protein interface in DTD-like fold. Curr Opin Struct Biol 53: 107-114. Link: https://bit.ly/2Y3XnSE
- Tamura T, Nakano S, Nakata E, Morii T (2017) Construction of a library of structurally diverse ribonucleopeptides with catalytic groups. Bioorg Med Chem 25: 1881-1888. Link: https://bit.ly/345K9IU
- Pichon C, Felden B (2007) Proteins that interact with bacterial small RNA regulators. FEMS Microbiol Rev 31: 614-625. Link: https://bit.ly/3h2UoBz
- Lin SS, Chang SC, Wang YH, Sun CY, Chang MF (2000) Specific interaction between the hepatitis delta virus RNA and glyceraldehyde 3-phosphate dehydrogenase: an enhancement on ribozyme catalysis. Virology 271: 46-57. Link: https://bit.ly/2CC8efc
- Arutyunova EI, Danshina PV, Domnina LV, Pleten AP, Muronetz VI (2003) Oxidation of glyceraldehyde-3-phosphate dehydrogenase enhances its binding to nucleic acids. Biochem Biophys Res Commun 307: 547-552. Link: https://bit.ly/3g5Ml5z
- Heinrich CP, Noack K, Wiss O (1972) Chemical modification of tryptophan at the binding site of thiamine-pyrophosphate in transketolase from Baker's yeast. Biochem Biophys Res Commun 49: 1427-1432. Link: https://bit.ly/2PXmmm2
- Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72: 248-254. Link: https://bit.ly/346zTjF
- Barbas CF, Burton DR, Scott JK, Silverman GJ (2007) Quantitation of DNA and RNA. Cold Spring Harb Protoc pdb.ip 47. Link: https://bit.ly/2E6cpjQ
- Schellenberger A, Hübner G (1965) On the separation of phosphoric acid esters of thiamine and its analogues by gradient elution. Hoppe Seylers Z Physiol Chem 343: 189-192. Link: https://bit.ly/345AmT7
- Datta AG, Racker E (1961) Mechanism of action of transketolase. II. The substrate–enzyme intermediate. J Biol Chem 236: 624-628. Link: https://bit.ly/2PZuRx3
- Kochetov GA (1982) Transketolase from yeast, rat liver, and pig liver. Methods Enzymol 90: 209-223. Link: https://bit.ly/3432cQ1
- Racker E, de Ia Haba G, Leder JG (1955) Crystalline transketolase from baker’s yeast: isolation and properties. J Biol Chem 214: 409-426. Link: https://bit.ly/2EcJt9z
- Kochetov GA, Philippov PP (1970) Calcium: cofactor of transketolase from baker's yeast. Biochem Biophys Res Commun 38: 930-933. Link: https://bit.ly/2Cz03A9
- Solovjeva ON, Selivanov VA, Orlov VN, Kochetov GA (2019) Stages of the formation of nonequivalence of active centers of transketolase from baker’s yeast. Mol Catal 466: 122-129. Link: https://bit.ly/3iQnswG
- Beck WS (1956) The Determination of triosephosphate isomerase. Arch Biochem Biophys 60: l-6. Link: https://bit.ly/2Cyv8Ed
- Dische Z, Dische R (1958) Methods for the qualitative and quantitative determinations of tetroses by two new specific color reactions. Biochim Biophys Acta 27: 184-188. Link: https://bit.ly/2Y91hK3
- Norton IL, Hartman FC (1972) Haloacetol phosphates. A comparative study of the active sites of yeast and muscle triose phosphate isomerase. Biochemistry 11: 4435-4441. Link: https://bit.ly/3g5Ln9r
- Nilson U, Hecquet L, Gefflaut T, Guerard C, Schneider G (1998) Asp477 is a determinant of the enantioselectivity in yeast transketolase. FEBS Lett 424: 49–52. Link: https://bit.ly/3aypWfS
- Bykova IA, Solovjeva ON, Meshalkina LE, Kovina MV, Kochetov GA (2001) One-substrate transketolase-catalyzed reaction. Biochem Biophys Res Commun 280: 845-847.Link: https://bit.ly/2Y64l9I
- Krietsch WK, Pentchevh PG, Klingenbürg H, Hofstätter T, Bücher T (1970) The isolation and crystallization of yeast and rabbit liver triose phosphate isomerase and a comparative characterization with the rabbit muscle enzyme. Eur J Biochem 14: 289-300. Link: https://bit.ly/3iT4gyl
- Vizán P, Alcarraz-Vizán G, Díaz-Moralli S, Solovjeva ON, Frederiks WM, et al. (2009) Modulation of pentose phosphate pathway during cell cycle progression in human colon adenocarcinoma cell line HT29. Int J Cancer 124: 2789-2796. Link: https://bit.ly/2FxvvA3
- Benito A, Polat IH, Noé V, Ciudad CJ, Marin S, et al. (2017) Glucose-6-phosphate dehydrogenase and transketolase modulate breast cancer cell metabolic reprogramming and correlate with poor patient outcome. Oncotarget 8: 106693-106706.Link: https://bit.ly/3237ice

Article Alerts
Subscribe to our articles alerts and stay tuned.
 This work is licensed under a Creative Commons Attribution 4.0 International License.
This work is licensed under a Creative Commons Attribution 4.0 International License.
 Save to Mendeley
Save to Mendeley
