Interaction of 6-Bromo- and 6-Chloro-Ubiquinone derivatives with mitochondrial electron transfer system
1Department of Biochemistry and Molecular Biology, Oklahoma State University, Stillwater, Oklahoma, 74078, USA
2Department of Chemistry, Tsinghua University, Beijing 100084, P.R. China
3School of Pharmaceutical Sciences, Sun Yat-sen University, Guangzhou, University City, Guangzhou 510006, China
Author and article information
Cite this as
Yu CA, Li Xl, Gu LQ, Yu L (2024) Interaction of 6-Bromo- and 6-Chloro-Ubiquinone derivatives with mitochondrial electron transfer system. Open J Chem. 2024; 10(1): 024-031. Available from: 10.17352/ojc.000036
Copyright License
© 2024 Yu CA, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.To understand the reaction mechanism of quinone-mediated electron transfer, a series of ubiquinone (Q)-derivatives with a bromine or chlorine atom at the 6-position and a different alkyl side chains at the 5-position of the benzoquinone ring were synthesized and characterized. The chemical properties and electron transfer activities were compared with the native ubiquinone, Q0C10. The redox midpoint potential of 6-Bromo- and 6-chloro-Q derivatives is 142 mV and 148 mV, respectively. These 6-halo-Q derivatives are reducible by ascorbate alone or by succinate in the presence of succinate-ubiquinone reductase. The reduced 6-halo-Q (6-halo-QH2) can be oxidized by cytochrome c alone or via ubiquinol-cytochrome c reductase. In the presence of cytochrome c oxidase and cytochrome c, 6-halo-QH2 is oxidizable by oxygen. 6-Halo-Q derivatives are reducible by succinate via succinate-ubiquinone reductase. They function as an electron-mediator connecting succinate-Q reductase and cytochrome c oxidase and bypass ubiquinol-cytochrome c reductase in the mitochondrial electron-transfer chain. This bypassing results in a decrease in energy coupling efficiency and a lower P/O ratio. These compounds might have therapeutic value in treating diseases that are caused by the oversupply of energy.
CcO: Cytochrome c Oxidase; Q0: 2,3-dimethoxy-5-methyl- 1,4-benzoquinone; Q0C10: 2,3-dimethoxy-5-decyl-6-methyl-1,4-benzoquinone; QCR: Ubiquinol- Cytochrome c Reductase; SCR: Succinate-Cytochrome c Oxidoreductase; SQR: Succinate-Ubiquinone Oxidoreductase. NADH: Reduced form of Nicotinamide Adenine Dinucleotide.
Introduction
The participation of ubiquinone (Q) in the mitochondrial electron-transfer chain has been well established [1-3]. However, the interaction between Q and protein and the reaction mechanism of Q-mediated electron transfer is not yet fully understood. The relative abundance of Q compared to other redox components in the inner mitochondrial membrane, along with its strong lipophilic nature, has led investigators to propose that Q functions as a homogeneous mobile carrier that shuttles electrons between complexes [4]. Several dynamic and kinetic studies support the idea that ubiquinone has a pool function in the mitochondrial electron transport system [5]. On the other hand, some studies indicate heterogeneity in the Q population of the inner mitochondrial membrane. The identification of the Q-binding proteins (site) in mitochondrial NADH-ubiquinone reductase (NQR) [6,7], succinate-ubiquinone reductase (SQR), and ubiquinol-cytochrome c reductase (QCR) support the idea that a Q-protein complex is an active specie during electron transfer [8]. Both schools of thought regarding the reaction mechanism of Q acknowledge the presence of specific Q-binding sites in electron transfer complexes. Q binding sites are clearly established by X-ray crystallographic studies in QCR [9-12], SQR [13], and quinol-fumarate reductase [14-16]. Also helping to establish quinone binding sites are photo-affinity labeling [17] and molecular genetic [18] approaches in SQR and QCR. Although the atomic structure of NQR is only available recently [19,20], the Q binding in the complex has been studied by photoaffinity labeling and inhibitor binding studies [21,22].
One of the approaches to studying Q-protein interactions and Q-binding sites of the Q-mediated electron transfer complexes is to elucidate the structural requirements for Q. This is aCcOmplished by using various synthetic Q-derivatives with different substituents on the benzoquinone ring [23] and various alkyl side chains [24]. The structural requirements for Q to serve as an electron donor or acceptor differ significantly. When Q is used as an electron acceptor for SQR, an alkyl side chain of six or more carbons gives maximum activity; when Q is used as an electron donor for QCR, an alkyl side chain of ten carbons gives maximum activity [25]. When Q is used as an electron acceptor for SQR, the methyl group at the 6-position is less important than the methoxy group at the 2- or 3-position [23]. When the electron-accepting activities of Q-derivatives with only one methoxy group are compared, the one with methoxy at the 3-position is more active than the one with methoxy at the 2-position [23]. The structural requirements for substituents on the benzoquinone ring are less strict when Q serves as an electron donor for QCR.
The use of chemically modified Q to study the interaction between Q and electron transfer protein complex has been well entertained since the 1980s when many modified Q became available [25]. A modified Q known as Q0C10OH was synthesized for the preparation of fluorescence- and photoaffinity-label Q in 1982. Q0C10OH was later developed into a successful cardiac drug named idebenone [26].
Continuing our efforts to understand the reaction mechanism of quinone-mediated electron transfer, a series of Q-derivative with a bromine or chlorine atom at the 6-position and a different alkyl side chain at the 5-position of a benzoquinone ring [6-bromo-2,3-dimethoxy-5-alkyl-1,4-benzoquinone and 6-chloro-2,3-dimethoxy-6-alkyl-1,4-benzoquinone] were synthesized and their chemical properties and electron transfer activities were compared to Q0C10. The non-enzymatic interaction of these derivatives with cytochrome c and their effect on the energy coupling efficiency of mitochondria were also investigated.
Experimental procedures
Materials
Cytochrome c, Type III, and 2, 3-dimethoxy-5-methyl-1, 4-benzoquinone (Q0) were purchased from Sigma; silica gel G thin layer plates from Analtech; 2, 3-dimethoxy-5-methyl-6-decyl-1, 4-benzoquinone (Q0C10) were synthesized as previously reported [25]. Azolectin (crude soybean phospholipids) was a product of Associate Concentrate and it was partially purified aCcOrding to Sone, et al. [27]. Other chemicals were of the highest purity that were commercially available.
Enzyme preparations
Succinate-ubiquinone reductase (SQR) [28], succinate-cytochrome c reductase (SCR) [28], QCR [29], and cytochrome c oxidase (CcO) [30] were prepared from beef heart submitochondrial particles (SMP) and assayed aCcOrding to the reported methods. The preparation of SQR was dispersed in 50 mM phosphate buffer, pH 8.0, containing 0.2% sodium cholate. The preparation of QCR was dispersed in 50 mM Tris-HCl buffer, pH 8.0, containing 0.66 M sucrose and 0.01 % dodecyl maltoside. The preparation of the CcO was dispersed in 50 mM phosphate buffer, pH 7.4, containing 1.5% sodium cholate.
Intact mitochondria were prepared from fresh rat liver and assayed by reported methods [31]. The consummation of oxygen for mitochondria was measured by YSI Model 53 Biological Oxygen Monitor at room temperature while stirring.
The concentrations of cytochromes b and c1 were determined spectrophotometrically, using difference millimolar coefficients of 28.5 and 17.5 for reduced minus oxidized absorbance at 562 nm minus 575 nm and at 552 nm minus 540 nm for cytochromes b [32] and c1 [32] respectively. Sodium dithionite and ascorbate were used as the reductants for cytochromes b and c1, respectively.
Protein assays were performed aCcOrding to the Lowry method in the presence of 1% SDS, with crystalline bovine serum albumin as standard [33].
Spectral Measurements
Absorption spectra were measured in a Shimadzu spectrophotometer model 2101PC. 1H NMR spectra were measured in a Varian XL-400 NMR spectrometer. The molecular weights of the Q derivatives were determined with a V6 ZAB-2SE high-resolution mass spectrometer.
Proton pumping assay for QCR vesicles
QCR was incorporated into azolectin phospholipid vesicles (liposomes) by cholate dialysis method [34]. The bulk-phase proton gradient generated by protein-PL vesicles was determined with a pH electrode (for outward proton ejection). For pH measurements, a Beckman electrode model 39849 and an Accument pH meter model 10 (Fisher Scientific) were used.
Redox potentials and relative hydrophobicity
The redox potentials of Q derivatives were determined from the ratio of reduced and oxidized forms of Q0 and Q0H2 after a short exposure to pH 12. They were then neutralized with acid under anaerobic conditions. Reduced Q0 was prepared by the treatment of NaBH4 followed by hexane extraction. The buffer system used was 50 % ethanol in 50 mM phosphate pH 7.0. A midpoint potential of 110 mV was used for Q0 in the calculations. The relative hydrophobicity of Q derivatives was determined by their Rfs in the thin layer chromatography, using a hexane/ether (3:1) mixture as a developing solvent system.
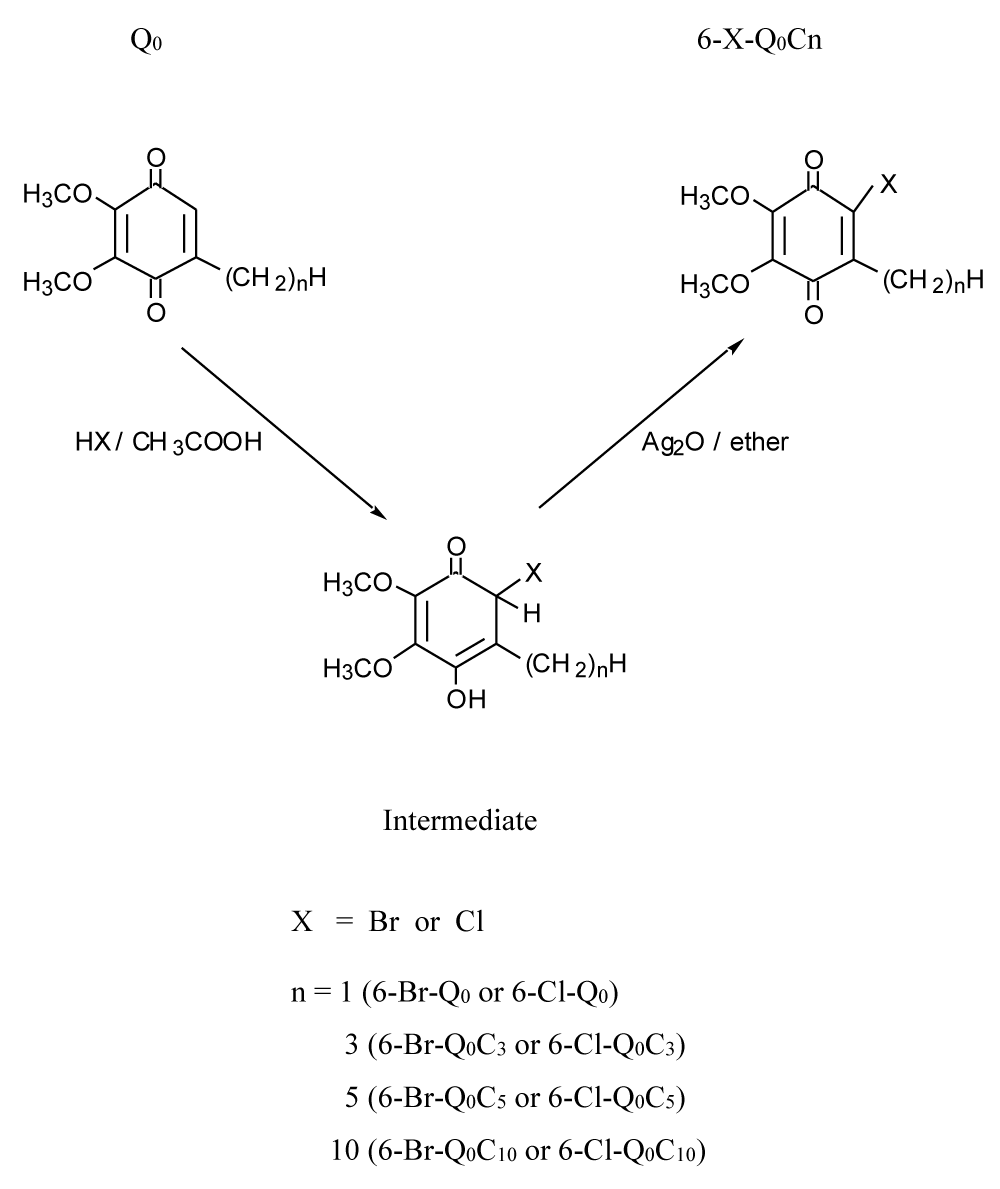
Synthesis of 6-bromo-2, 3-dimethoxy-5-alkyl-1, 4-benzoquinones and 6-chloro-2, 3-dimethoxy-5-alkyl-1, 4-benzoquinones: Figure 1 summarized the method used in the synthesis of those 6-bromo- and 6-chloro-Q derivatives.
6-Bromo-2, 3-dimethoxy-5-methyl-1, 4-benzoquinone (6-Br-Q0): One hundred and eighty mg of Q0 (2,3-dimethoxy-5-methyl-1,4-benzoquinone, 1 mmol) in 5 mL of acetic acid was mixed with 5 ml of 48% hydrobromic acid. The mixture was stirred at room temperature for 30 minutes, then diluted with 40 mL of water and extracted with ether. The ether extract was washed with H2O and dried over anhydrous Na2SO4. One gram of Ag2O was added to the dried ether solution and the mixture was stirred for one hour at room temperature. The solid phase was removed by filtration, and the filtrate solution was dried in a rotary evaporator. The crude product was purified by TLC (solvent system: ether/hexane mixtures), yielding 92%. 6-Br-Q0 is a red crystalline, m. p. 58-60°C. H NMR (CDCl3): 2.20 (s, 3H), 4.02 (s, 3H), 4.06 (s, 3H) ppm. UVEtOH: oxid, 302 nm; red, 293 nm. MS (m/z), 260 (M), 262 (M+2); HRMS, 259.9681 (cal. 259.9684).
By using the same method, a series of 6-bromo- and 6-chloro-Q derivatives were synthesized. Starting materials, such as 2,3-dimethoxy-1,4-benzoquinone, 2,3-dimethoxy-5-propy-1,4-benzoquinone, 2,3-dimethoxy-5-pentyl-1,4-benzoquinone, and 2,3-dimethoxy-5-decyl-1,4-benzoquinone, were prepared aCcOrding to the methods described previously.
6-Bromo-2, 3-dimethoxy-5-propyl-1, 4-benzoquinone (6-Br-Q0C3): Red oil. 1H NMR (CDCl3): 1.02 (t, 3H), 1.75 (m, 2H), 2.20 (t, 2H), 4.02 (s, 3H), 4.06 (s, 3H) ppm. UVEtOH: oxid, 302 nm; red, 293 nm. MS (m/z): 288 (M); 290 (M+2).
6-Bromo-2, 3-dimethoxy-5-pentyl-1, 4-benzoquinone (6-Br-Q0C5): Red oil. 1H NMR (CDCl3): 0.89 (t, 3H), 1.28-1.75 (m, 6H), 2.20 (t, 2H), 4.02 (s, 3H), 4.06 (s, 3H) ppm. UVEtOH: oxid, 302 nm; red, 293 nm. MS (m/z): 316 (M); 318 (M+2).
6-Brom-2, 3-dimethoxy-5-decyl-1, 4-benzoquinone (6-Br-Q0C10): Red oil. 1H NMR (CDCl3): 0.89 (t, 3H), 1.23-1.6 (m, 16H) 2.20 (t, 2H), 4.02 (s, 3H), 4.06 (s, 3H) ppm. UVEtOH: oxid, 302 nm; red, 293 nm. MS (m/z): 386 (M); 388 (M+2); HRMS, 386.1098 (cal. 386.1093).
6-Chloro-2, 3-dimethoxy-5-methyl-1,4-benzoquinone (6-Cl-Q0): Red crystalline. 1H NMR (CDCl3): 2.18 (s, 3H), 4.01 (s, 3H), 4.05 (s, 3H) ppm. UVEtOH: oxide, 287 nm; red, 290 nm. MS(m/z), 216 (M), 218 (M+2); HRMS, 216.0185 (cal. 216.0190).
6-Chloro-2, 3-dimethoxy-5-propyl-1,4-benzoquinone (6-Cl-Q0C3): Red oil. 1H NMR (CDCl3): 0.95 (t, 3H), 1.62 (m, 2H) 2.16 (t, 2H), 4.01 (s, 3H), 4.04 (s, 3H) ppm. UVEtOH: oxid, 287 nm; red, 290 nm. MS (m/z): 244 (M); 246 (M+2).
6-Chloro-2, 3-dimethoxy-5-pentyl-1,4-benzoquinone (6-Cl-Q0C5): Red oil. 1H NMR (CDCl3): 0.89 (t, 3H), 1.20-1.62 (m, 6H), 2.15 (t, 2H), 4.01 (s, 3H), 4.04 (s, 3H) ppm. UVEtOH: oxid, 287 nm; red, 290 nm. MS (m/z): 272 (M); 274 (M+2).
6-Chloro-2, 3-dimethoxy-5-decyl-1, 4-benzoquinone (6-Cl-Q0C10): Red oil. 1H NMR (CDCl3): 0.89 (t, 3H), 1.18-1.60 (m, 16H), 2.14 (t, 2H), 4.01 (s, 3H), 4.04 (s, 3H) ppm. UVEtOH: oxid, 287 nm; red, 290 nm. MS (m/z): 342 (M); 344 (M+2).
Results and discussions
Structures and properties of 6-bromo- and 6-chloro-Q derivatives
Figure 1 shows the chemical structures of 6-bromo- and 6-chloro-Q derivatives. The absorption spectral properties of 6-bromo- and 6-chloro-Q are similar to those of 2,3-dimethoxy-5-decyl-6-methyl-1,4-benzoquinone (Q0C10) but differ respectively with 16 nm and 8 nm red shifts in the oxidized forms, and with 5 nm and 3 nm red shift in the reduced forms.
The midpoint potential (Em) of 6-bromo- and 6-chloro-Q derivatives were determined to be 142 mV and 148 mV, respectively. The introduction of an electron-withdrawing group such as a bromine or chlorine atom to the benzoquinone ring results in the stabilization of the reduced form of the derivatives and thus increases their redox midpoint potentials.
6-Bromo- and 6-chloro-Q derivatives as electron acceptors for succinate-ubiquinone reductase (SQR)
Table 1 summarizes the maximum electron acceptor activities of 6-bromo- and 6-chloro-Q derivatives as compared to that of Q0C10 from succinate catalyzed by SQR.
All the halo-Q derivatives show partial electron acceptor activities regardless of the presence or absence of the alkyl side chain at the 5-position of the benzoquinone ring. The electron acceptor activity increased slightly as the size of the 5-alkyl group increased.
Interaction of reduced 6-bromo and 6-chloro-Q derivatives with cytochrome c
Unlike other ubiquinone homologs which show little interaction with cytochrome c, 6-halo-Q derivatives with an alkyl side chain of less than five carbons interact with cytochrome c freely in the absence of an electron transfer protein complex. The equilibration is dependent on the redox states and concentrations. Since the midpoint redox potentials of 6-halo-Q derivatives are lower than that of cytochrome c, the following reaction is favored on the reduction of cytochrome c. As expected, the reaction is pH-dependent. At pH lower than 7, very little 6-halo-QH0 is oxidized by cytochrome c. This is because the midpoint potentials of the 6-halo-Q derivatives increased as the pH decreased.
6-X-QH2 + cyt. c 3+ → 6-X-Q + cyt. c 2+ + 2H+
X = Br or Cl
6-Halo-Q derivatives with an alkyl side chain of more than five carbons show little interaction with cytochrome c, but the interaction with cytochrome c increases significantly in the presence of QCR. Table 2 shows the interactions between 6-halo-Q derivatives and cytochrome c in the absence or presence of QCR.
Reduced 6-Bromo- and 6-chloro-Q derivatives as electron donors for ubiquinol-cytochrome c reductase
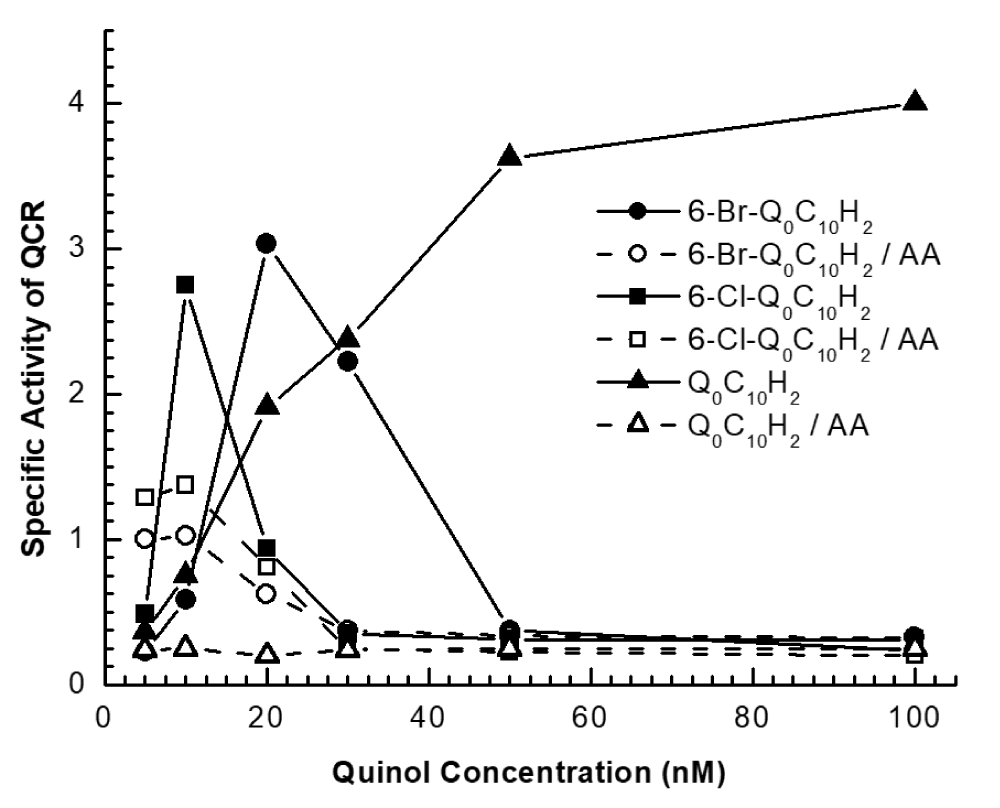
Table 2 summarizes the oxidation of reduced 6-halo-Q derivatives by cytochrome c in the presence or absence of QCR. Reduced 6-halo-Q derivatives with short alkyl side chains or no side chains can be oxidized by cytochrome c non-enzymatically. Their oxidizability decreases as the sizes of the alkyl side chain increase. The reduced 6-halo-Q derivatives with alkyl side chains can function as electron donors for QCR. The enzymatic oxidizability increases as the size of the alky side chain increases. The maximal oxidizability is reached with the derivative having ten carbons in the alkyl side chain.
Figure 2 shows the concentration-dependent oxidation of reduced 6-halo-Q derivatives by cytochrome c catalyzed by QCR. The activity increased with concentrations of 6-halo-Q derivatives increased to a maximum and then decreased. At a high concentration of reduced derivatives, very little oxidation was observed. This substrate inhibition is not observed in regular ubiquinol unless an extremely high concentration is used. The sharp decrease in oxidation of halo-Q derivatives is an interesting phenomenon that cannot be explained easily. One possibility is that with the high concentrations of reduced halo-Q derivatives, they compete with the oxidized form Q derivatives for the quinone reduction (Qi or QN) site of QCR and thus inhibit the activity. ACcOrding to the structural information of the QN site of QCR, ubiquinol is bound through two hydrogen bonds, one between the oxygen in 2-methoxy and the hydrogen of the hydroxyl of serine-205, and the other between 1-hydroxyl of ubiquinol and aspartate-228 bridged by a water molecule [35]. Near the methyl group of ubiquinol, there is lysine-227 [35], which may increase the binding of ubiquinol by having its methyl group replaced by a bromine or chlorine atom.
It should be mentioned that this enzymatic electron-transfer reaction is partially sensitive to antimycin A (Figure 2).
6-bromo- and 6-chloro-Q derivatives function as an electron shunt between SQR and cytochrome c oxidase (CcO) in the electron transfer chain
Since the 6-halo-Q derivatives are reducible by succinate in the presence of succinate-Q reductase, and the reduced derivatives are oxidized by cytochrome c directly in the absence of QCR. These derivatives are expected to function as an electron shunt between SQR and CcO complexes. In the presence of SQR and CcO, an electron is transferred from succinate to oxygen directly without involving QCR.
Succ. → SQR 6-X-Q → Cyt. c → CcO → O2
As expected, this electron transfer reaction is sensitive to CcO inhibitors, such as CO or KCN.
When purified QCR is embedded in phospholipid vesicles, it pumps protons out of the vesicles during the oxidation of ubiquinol (Q0C10H2). The vesicles also transfer an electron to cytochrome c with an H+/e- ratio of about 2. When reduced 6-halo-Q derivatives were used as substrate for cytochrome bc1 embedded vesicles, very little proton pumping activity was observed (see Table 3), even with the derivatives having a large alkyl side chain. The lack of proton pumping activity during the oxidation of reduced 6-halo-Q derivatives cannot be simply attributed to the non-enzymatic oxidation of derivatives by cytochrome c. Although derivatives with a large alkyl side chain have better enzymatic oxidation activity (80% - 95%) than the non-enzymatic one (5% - 20%), their proton pumping activities are still very low.
Table 4 summarizes the oxygen uptake rates in the electron transfer chain from SQR to CcO in the presence or absence of cytochrome c, using 6-halo-Q derivatives as an electron mediator.
The bypass ability of 6-halo-Q derivatives to QCR in the electron transfer chain is dependent on the carbon chain length of the alkyl group of the derivatives. Derivatives with an alkyl side chain of less than five carbons show significant bypass function. The compounds with an alkyl side chain of ten carbons have much less bypass activity.
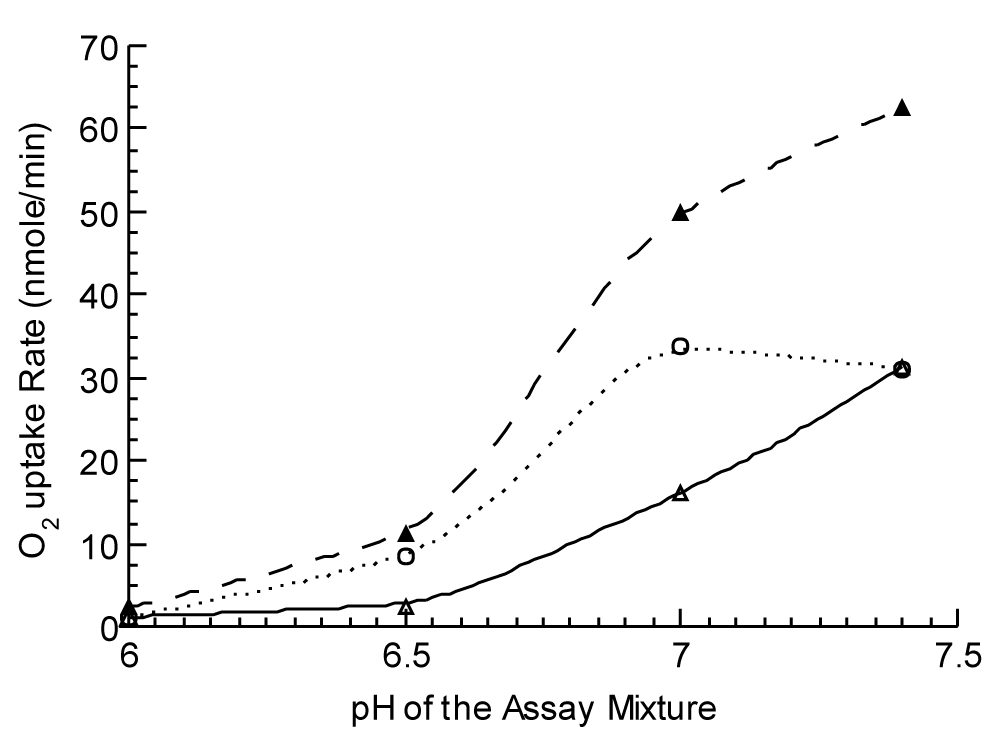
The observation that the 6-Br-Q0 and 6-Cl-Q0 have a much higher oxygen uptake rate than other derivatives with no halo-substituent such as Q0 is consistent with the results of the higher rates observed for the electron transfer from reduced Q to cytochrome c and the stronger interaction between reduced 6-halo-Q0H2 and oxygen (Table 4). We found that reduced form 6-halo-Q0H2 can be oxidized by oxygen in the absence or presence of CcO in the reaction mixture. As expected the oxidation in the absence of cytochrome c was not sensitive to cyanide. The interaction between reduced 6-halo-Q0 and oxygen is pH-dependent. Fig. 3 shows the effect of pH on the O2-uptake rate of SQR-reduced halo-Q derivatives.
These results indicate the electron transfer from reduced 6-Cl-Q0H2 to oxygen in the presence of CcO in the neutral pH range of 7.0 to 8.0 is a pH less sensitive, enzymatic reaction. For the reaction in the absence of CcO, (a non-enzymatic reaction), the oxidation rate increases as the pH of the reaction mixture increases.
The electrons transfer from 6-halo-Q0H2 to oxygen may take two pathways. One is via normal CcO and the reaction is cyanide sensitive. Another is that reduced 6-halo-Q derivatives directly interact with molecular oxygen and the reaction is cyanide insensitive.
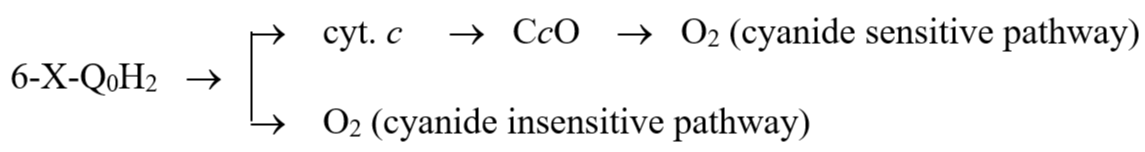
X = Br or Cl
Effects of 6-bromo-and 6-chloro-Q derivatives effect on state 3 respiration of mitochondria
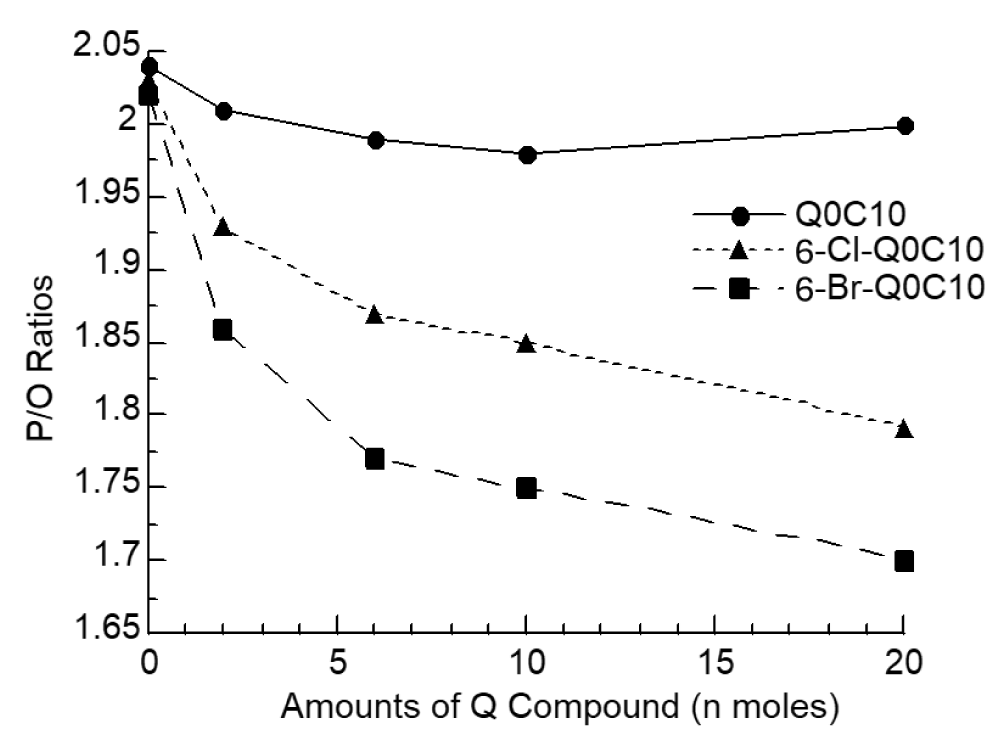
Unlike other homologs of ubiquinone, 6-halo-Q derivatives showed an inhibitory effect on the state 3 of NADH or succinate-supported respiration when the concentration is higher than 20 µM [34]. This concentration-dependent inhibition apparently results from the bypass of site 2 energy conservation. The electrons derived from NADH, or succinate, can bypass the QCR region of the respiratory chain to reduce oxygen. As a result, less energy is conserved leading to a decrease in stimulation of respiration upon the addition of ADP in the mitochondria. The P/Q ratio [36] decreased as the concentration of 6-halo-Q derivatives used increased (Figure 4).
Concluding remarks
More than 90% of the energy required to sustain life, growth, and the structural integrity of a living organism is derived from a process called oxidative phosphorylation. This process takes place in the mitochondrial inner membrane of eukaryotic cells or in the cytoplasmic membrane of prokaryotic organisms, via a coupling reaction of two multi-subunit membrane protein complexes: the electron transfer chain complexes Complexes I (NQR), II (SQR), III (QCR) and IV (CcO), and Complex V (ATP synthase complex). Coenzyme Q, which functions as an electron acceptor for NQR and SQ, and as an electron donor for QCR, is the only component whose concentration can be modified externally. The idea of using chemically synthesized Q to improve the bioenergetic function has been well entertained. The idea of using modified coenzyme Q to decrease the bioenergetic function, however, is relatively new. Taking advantage of the availability of 6-Br-Q0C10, we have recently investigated the effect of this compound on cell growth using commercially available cell lines and body weight gain in rats. As expected, our preliminary results show that the 6-Br-Q0C10 indeed reduces cell growth by 30-60% depending on the type of cells used. It decreases rat’s boy weight gain by 30% - 40%. These suggest that 6-Br-Q0C10 or 6-Cl- Q0C10 has a great potential to reduce obesity and slow the growth of cancer cells. A patent application based on these findings has been developed and submitted.
This work was supported by grants GM30721 from the National Institutes of Health, and the Oklahoma Agricultural Experiment Station (Projects #1819 and #2372), Oklahoma State University.
- Crane FL. Hydroquinone dehydrogenases. Annu Rev Biochem. 1977;46:439-69. doi: 10.1146/annurev.bi.46.070177.002255. PMID: 197879.
- Ernster L, Lee IY, Norling B, Persson B. Studies with ubiquinone-depleted submitochondrial particles. Essentiality of ubiquinone for the interaction of succinate dehydrogenase, NADH dehydrogenase, and cytochrome b. Eur J Biochem. 1969 Jun;9(3):299-310. doi: 10.1111/j.1432-1033.1969.tb00609.x. PMID: 4307591.
- Elgar K. Coenzyme Q10: A Review of Clinical Use and Efficacy. Nutr Med J. 2021; 1 (1): 100-118.
- Gupte S, Wu ES, Hoechli L, Hoechli M, Jacobson K, Sowers AE, Hackenbrock CR. Relationship between lateral diffusion, collision frequency, and electron transfer of mitochondrial inner membrane oxidation-reduction components. Proc Natl Acad Sci U S A. 1984 May;81(9):2606-10. doi: 10.1073/pnas.81.9.2606. PMID: 6326133; PMCID: PMC345118.
- Kröger A, Klingenberg M. The kinetics of the redox reactions of ubiquinone related to the electron-transport activity in the respiratory chain. Eur J Biochem. 1973 Apr;34(2):358-68. doi: 10.1111/j.1432-1033.1973.tb02767.x. PMID: 4351161.
- Xu Z, Huo J, Ding X, Yang M, Li L, Dai J, Hosoe K, Kubo H, Mori M, Higuchi K, Sawashita J. Coenzyme Q10 Improves Lipid Metabolism and Ameliorates Obesity by Regulating CaMKII-Mediated PDE4 Inhibition. Sci Rep. 2017 Aug 15;7(1):8253. doi: 10.1038/s41598-017-08899-7. PMID: 28811612; PMCID: PMC5557856.
- Ragan CI, Cottingham IR. The kinetics of quinone pools in electron transport. Biochim Biophys Acta. 1985 Apr 8;811(1):13-31. doi: 10.1016/0304-4173(85)90003-5. PMID: 3986195.
- Yu CA, Yu L. Ubiquinone-binding proteins. Biochim Biophys Acta. 1981 Dec 4;639(2):99-128. doi: 10.1016/0304-4173(81)90007-0. PMID: 6272848.
- Xia D, Yu CA, Kim H, Xia JZ, Kachurin AM, Zhang L, Yu L, Deisenhofer J. Crystal structure of the cytochrome bc1 complex from bovine heart mitochondria. Science. 1997 Jul 4;277(5322):60-6. doi: 10.1126/science.277.5322.60. Erratum in: Science 1997 Dec 19;278(5346):2037. PMID: 9204897.
- Zhang Z, Huang L, Shulmeister VM, Chi YI, Kim KK, Hung LW, Crofts AR, Berry EA, Kim SH. Electron transfer by domain movement in cytochrome bc1. Nature. 1998 Apr 16;392(6677):677-84. doi: 10.1038/33612. PMID: 9565029.
- Iwata S, Lee JW, Okada K, Lee JK, Iwata M, Rasmussen B, Link TA, Ramaswamy S, Jap BK. Complete structure of the 11-subunit bovine mitochondrial cytochrome bc1 complex. Science. 1998 Jul 3;281(5373):64-71. doi: 10.1126/science.281.5373.64. PMID: 9651245.
- Hunte C, Koepke J, Lange C, Rossmanith T, Michel H. Structure at 2.3 A resolution of the cytochrome bc(1) complex from the yeast Saccharomyces cerevisiae co-crystallized with an antibody Fv fragment. Structure. 2000 Jun 15;8(6):669-84. doi: 10.1016/s0969-2126(00)00152-0. PMID: 10873857.
- Sun F, Huo X, Zhai Y, Wang A, Xu J, Su D, Bartlam M, Rao Z. Crystal structure of mitochondrial respiratory membrane protein complex II. Cell. 2005 Jul 1;121(7):1043-57. doi: 10.1016/j.cell.2005.05.025. PMID: 15989954.
- Iverson TM, Luna-Chavez C, Cecchini G, Rees DC. Structure of the Escherichia coli fumarate reductase respiratory complex. Science. 1999 Jun 18;284(5422):1961-6. doi: 10.1126/science.284.5422.1961. PMID: 10373108.
- Lancaster CR, Kröger A, Auer M, Michel H. Structure of fumarate reductase from Wolinella succinogenes at 2.2 A resolution. Nature. 1999 Nov 25;402(6760):377-85. doi: 10.1038/46483. PMID: 10586875.
- Iverson TM, Luna-Chavez C, Croal LR, Cecchini G, Rees DC. Crystallographic studies of the Escherichia coli quinol-fumarate reductase with inhibitors bound to the quinol-binding site. J Biol Chem. 2002 May 3;277(18):16124-30. doi: 10.1074/jbc.M200815200. Epub 2002 Feb 15. PMID: 11850430.
- Lee GY, He DY, Yu L, Yu CA. Identification of the ubiquinone-binding domain in QPs1 of succinate-ubiquinone reductase. J Biol Chem. 1995 Mar 17;270(11):6193-8. doi: 10.1074/jbc.270.11.6193. PMID: 7890754.
- Shenoy SK, Yu L, Yu Ca. Identification of quinone-binding and heme-ligating residues of the smallest membrane-anchoring subunit (QPs3) of bovine heart mitochondrial succinate:ubiquinone reductase. J Biol Chem. 1999 Mar 26;274(13):8717-22. doi: 10.1074/jbc.274.13.8717. PMID: 10085111.
- Efremov RG, Baradaran R, Sazanov LA. The architecture of respiratory complex I. Nature. 2010 May 27;465(7297):441-5. doi: 10.1038/nature09066. PMID: 20505720.
- Hunte C, Zickermann V, Brandt U. Functional modules and structural basis of conformational coupling in mitochondrial complex I. Science. 2010 Jul 23;329(5990):448-51. doi: 10.1126/science.1191046. Epub 2010 Jul 1. PMID: 20595580.
- Gong X, Xie T, Yu L, Hesterberg M, Scheide D, Friedrich T, Yu CA. The ubiquinone-binding site in NADH:ubiquinone oxidoreductase from Escherichia coli. J Biol Chem. 2003 Jul 11;278(28):25731-7. doi: 10.1074/jbc.M302361200. Epub 2003 May 2. PMID: 12730198.
- Nakamaru-Ogiso E, Sakamoto K, Matsuno-Yagi A, Miyoshi H, Yagi T. The ND5 subunit was labeled by a photoaffinity analogue of fenpyroximate in bovine mitochondrial complex I. Biochemistry. 2003 Jan 28;42(3):746-54. doi: 10.1021/bi0269660. PMID: 12534287.
- Gu LQ, Yu L, Yu CA. Effect of substituents of the benzoquinone ring on electron-transfer activities of ubiquinone derivatives. Biochim Biophys Acta. 1990 Feb 22;1015(3):482-92. doi: 10.1016/0005-2728(90)90082-f. PMID: 2154255.
- Yu CA, Gu LQ, Lin YZ, Yu L. Effect of alkyl side chain variation on the electron-transfer activity of ubiquinone derivatives. Biochemistry. 1985 Jul 16;24(15):3897-902. doi: 10.1021/bi00336a013. PMID: 2996584.
- Yu CA, Yu L. Syntheses of biologically active ubiquinone derivatives. Biochemistry. 1982 Aug 17;21(17):4096-101. doi: 10.1021/bi00260a028. PMID: 6289870.
- Wang J, Li S, Yang T, Yang J. Single-step synthesis of idebenone from Coenzyme Q0 via free-radical alkylation under silver catalysis. Tetrahedron. 2014; 70: 9029-9032.
- Sone N, Yoshida M, Hirata H, Kagawa Y. Reconstitution of vesicles capable of energy transformation from phospholipids and adenosine triphosphatase of a thermophilic bacterium. J Biochem. 1977 Feb;81(2):519-28. doi: 10.1093/oxfordjournals.jbchem.a131485. PMID: 14954.
- Yu L, Yu CA. Quantitative resolution of succinate-cytochrome c reductase into succinate-ubiquinone and ubiquinol-cytochrome c reductases. J Biol Chem. 1982 Feb 25;257(4):2016-21. PMID: 6276404.
- Yu L, Yang S, Yin Y, Cen X, Zhou F, Xia D, Yu CA. Chapter 25 Analysis of electron transfer and superoxide generation in the cytochrome bc1 complex. Methods Enzymol. 2009;456:459-73. doi: 10.1016/S0076-6879(08)04425-X. PMID: 19348904; PMCID: PMC7423196.
- Yu C, Yu L, King TE. Studies on cytochrome oxidase. Interactions of the cytochrome oxidase protein with phospholipids and cytochrome c. J Biol Chem. 1975 Feb 25;250(4):1383-92. PMID: 163252.
- Harmon HJ, Crane FL. Inhibition of mitochondrial electron transport by hydrophilic metal chelators. Determination of dehydrogenase topography. Biochim Biophys Acta. 1976 Jul 9;440(1):45-58. doi: 10.1016/0005-2728(76)90112-2. PMID: 947364.
- Berden JA, Slater EC. The reaction of antimycin with a cytochrome b preparation active in reconstitution of the respiratory chain. Biochim Biophys Acta. 1970 Sep 1;216(2):237-49. doi: 10.1016/0005-2728(70)90215-x. PMID: 5504626.
- LOWRY OH, ROSEBROUGH NJ, FARR AL, RANDALL RJ. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951 Nov;193(1):265-75. PMID: 14907713.
- Racker E, Kandrach A. Partial resolution of the enzymes catalyzing oxidative phosphorylation. XXXIX. Reconstitution of the third segment of oxidative phosphorylation. J Biol Chem. 1973 Aug 25;248(16):5841-7. PMID: 4353278.
- Xia D, Esser L, Tang WK, Zhou F, Zhou Y, Yu L, Yu CA. Structural analysis of cytochrome bc1 complexes: implications to the mechanism of function. Biochim Biophys Acta. 2013 Nov-Dec;1827(11-12):1278-94. doi: 10.1016/j.bbabio.2012.11.008. Epub 2012 Nov 29. PMID: 23201476; PMCID: PMC3593749.
- Vercesi A, Reynafarje B, Lehninger AL. Stoichiometry of H+ ejection and Ca2+ uptake coupled to electron transport in rat heart mitochondria. J Biol Chem. 1978 Sep 25;253(18):6379-85. PMID: 210182.
Article Alerts
Subscribe to our articles alerts and stay tuned.
 This work is licensed under a Creative Commons Attribution 4.0 International License.
This work is licensed under a Creative Commons Attribution 4.0 International License.


 Save to Mendeley
Save to Mendeley
