Conceptual Study of Enzymatic Organic Reactions in the Bacteriophage Therapy
1Organic Chemistry Professor, Department of Chemistry, Faculty of Science, University of Lubumbashi, Democratic Republic of Congo
2Research Associate at Hydro-Quebec Institute for Environment, Development and Society, Laval University, Quebec, Quebec, Canada
Author and article information
Cite this as
Mwene-Mbeja TM. Conceptual Study of Enzymatic Organic Reactions in the Bacteriophage Therapy. Open J Chem. 2024; 10(1): 073-079. Available from: 10.17352/ojc.000042
Copyright License
© 2024 Mwene-Mbeja TM. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.In order to neutralize bacteria, bacteriophages hydrolyze chemical constituents of the bacterial cellular membrane, such as phospholipids, glycolipids as well as glycoproteins before reaching the bacterial cytoplasm, an intracellular environment containing water, salts, and a diversity of organic compounds notably ribonucleic acid (RNA) along with deoxyribonucleic acid (DNA). In this perspective, when bacteriophages access the bacterial cytoplasm, they develop and consequently the excessive accumulation of bacteriophages facilitates or smooths the way for bacterial degradation.
This article aims to study the chemical behaviour of organic substances so as to better understand how bacteriophages destroy pathogen bacteria. In other words, bacteriophage therapy could be elucidated by applying enzymatic organic reactions knowledge. Indeed, when bacteriophages invade bacteria, they react with organic substances of bacterial cellular membranes such as phospholipids, glycolipids as well as glycoproteins. They also react with Ribonucleic Acid (RNA) and Deoxyribonucleic Acid (DNA) into the bacterial cytoplasm. Recall that organic substances have functional groups that act as acids, bases, electrophiles, or nucleophiles according to reaction conditions. Indeed, it should be mentioned that this article is innovative because it is the first time to display detailed organic reaction mechanisms so as to enhance comprehension of organic chemistry significance in bacteriophage therapy, compared to the bacteriophage action mechanisms published in the literature which do not reveal comprehensive advanced organic reactions mechanisms [1-6].
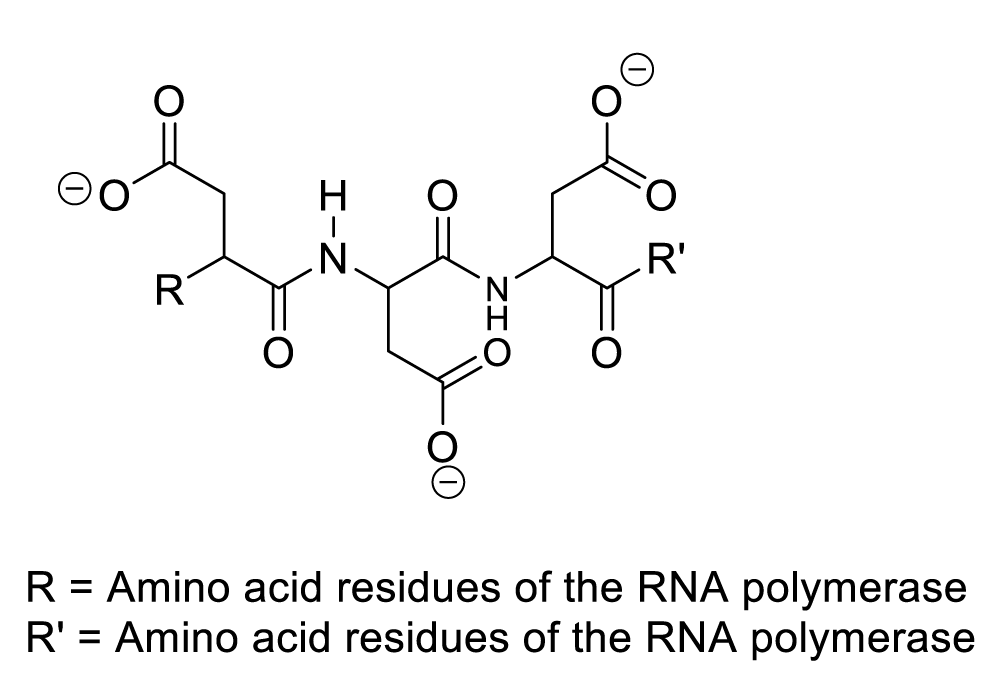
Bacteriophage therapy is a medical expertise that consists of eradicating infectious bacteria employing phages or bacteriophages. The latter are viruses containing ribonucleic acid or deoxyribonucleic acid wrapped by a protein and a catalytic centre of RNA polymerase [1-10]. Experimental observation has shown that three aspartate residues constitute the root of the catalytic centre of RNA polymerase (Figure 1) [1-10].
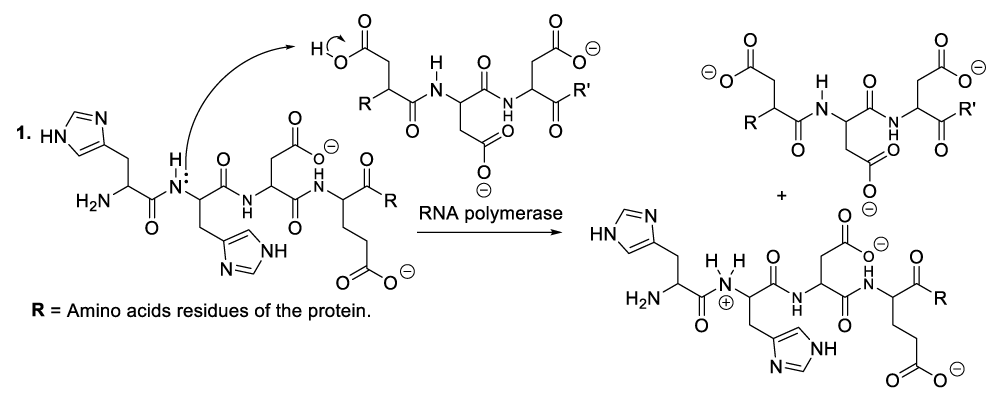
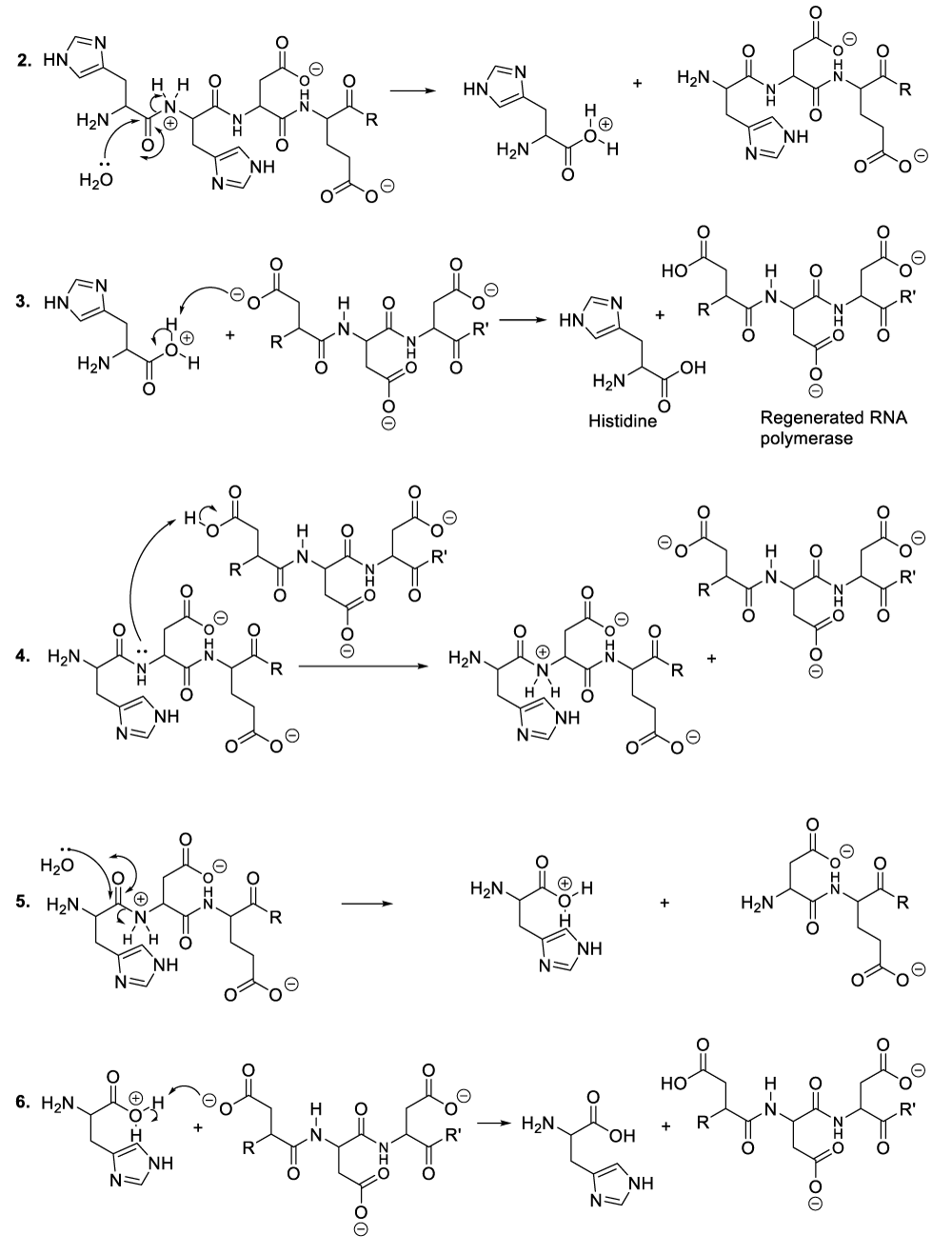
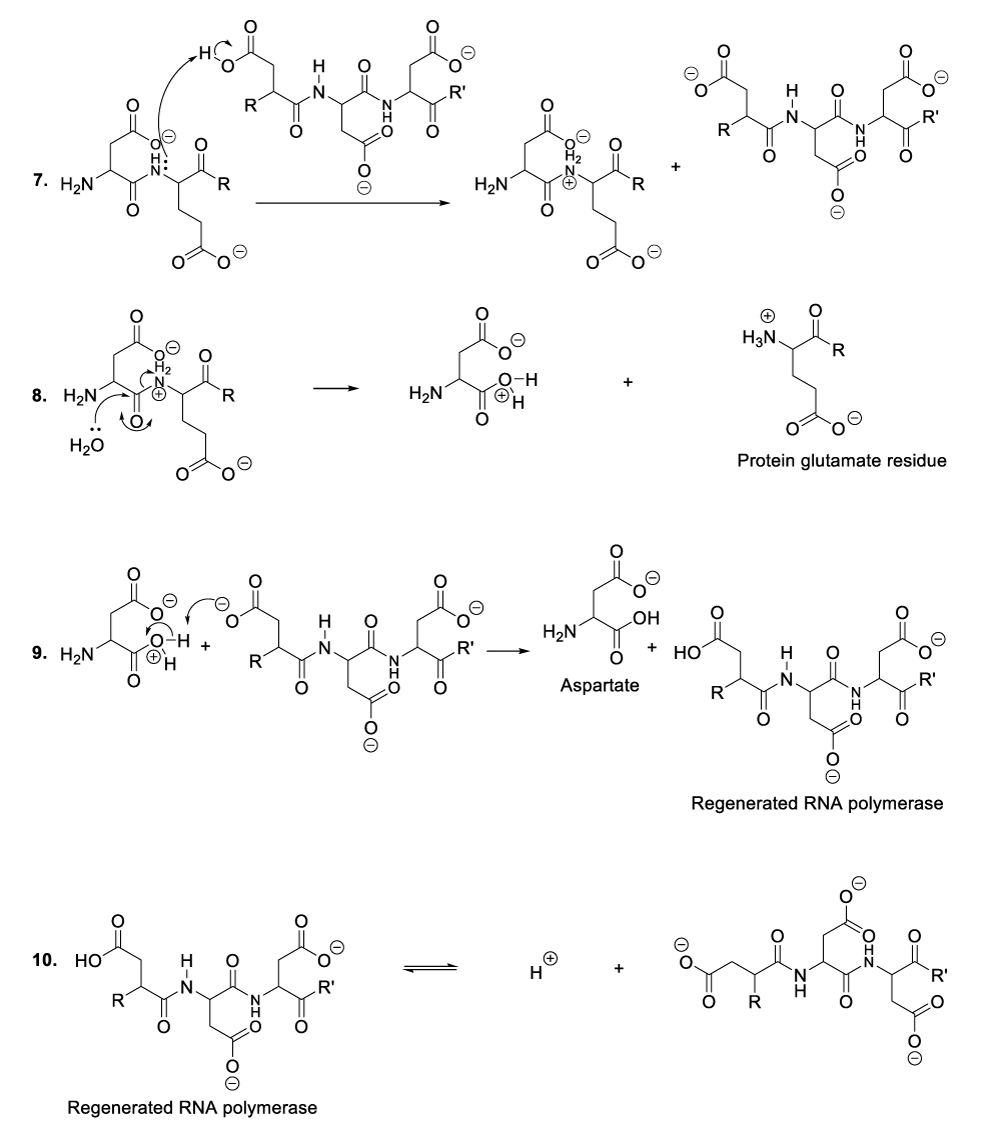
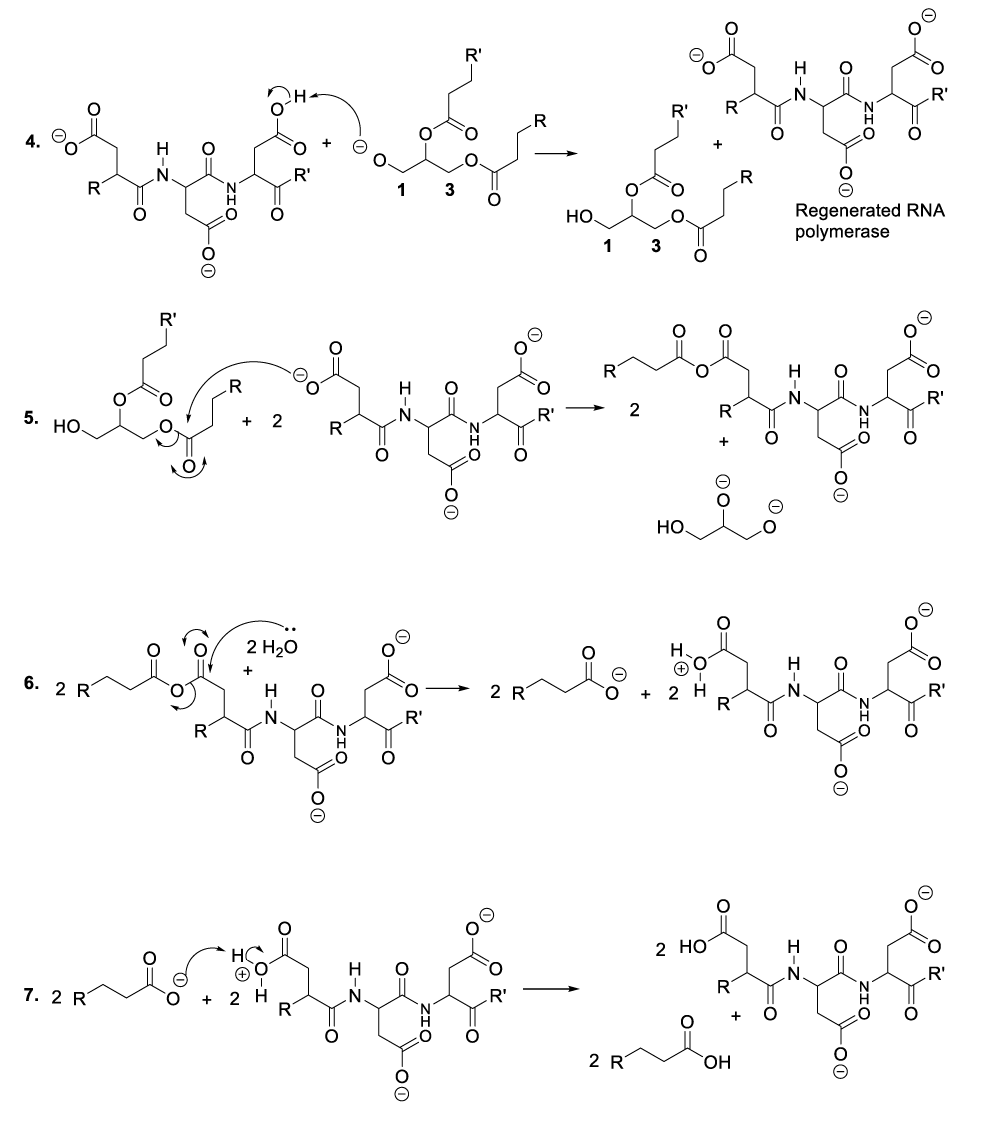
It is important to mention that the role of protein is to provide nutritional substances or amino acids to bacteriophages so that they can satisfy their nutritional needs, while they use aspartate residues to catalyze organic reactions (Scheme 1). In this context, enzymatic organic reactions demonstrate better the degradation of bacterial cellular membranes, and in such a circumstance, bacteriophages can easily reach bacterial cytoplasm where they spread for the purpose of destroying specific bacteria. In other words, the enzymatic organic reactions, which take place in the extracellular environment, involve the bacterial membrane together with the bacteriophage ARN polymerase. This aforementioned enzyme is capable of catalyzing hydrolysis of phospholipids, glycolipids, and glycoproteins that constitute the bacterial membrane. In the same perspective, the enzymatic organic reactions, which occur in the bacterial intracellular area, refer to the synthesis of many molecules of viral RNA or DNA (Scheme 5, Scheme 6).
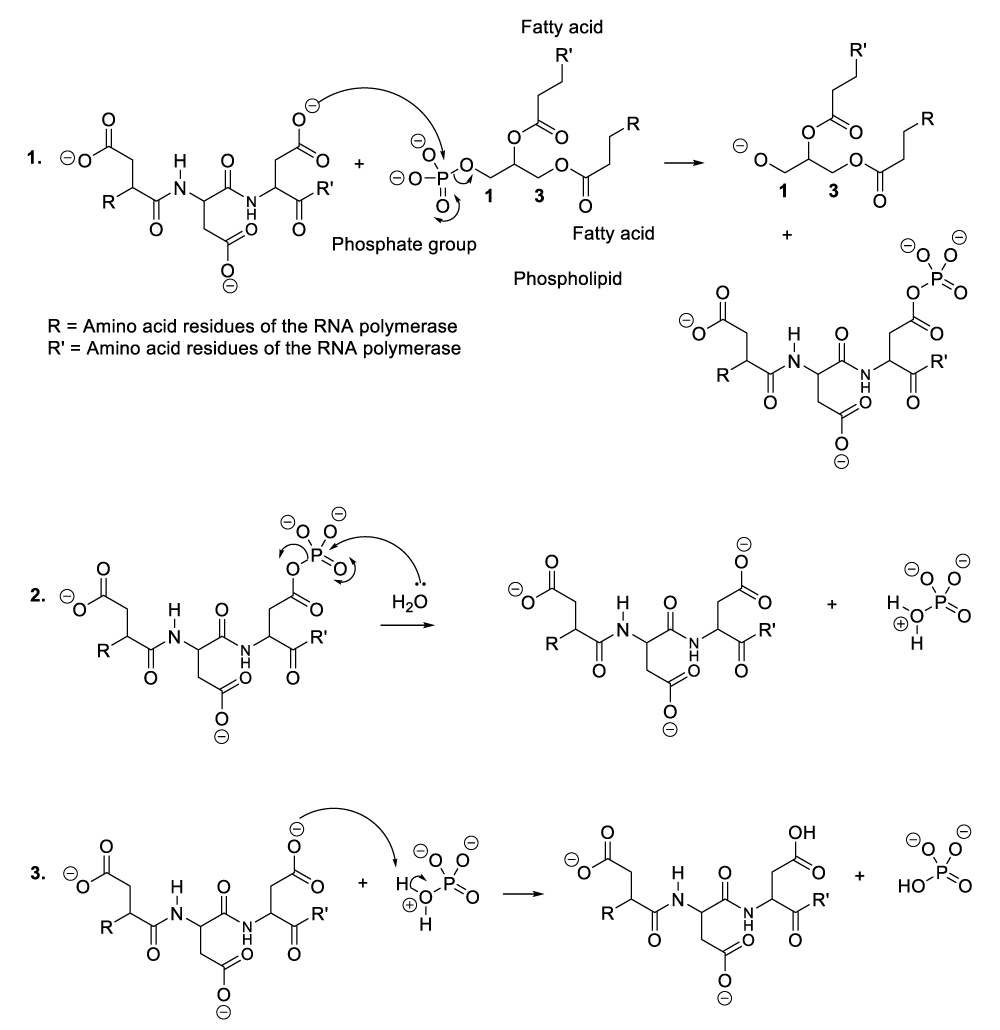
Enzymatic hydrolysis of phospholipids
A plausible explanation of the hydrolysis of phospholipids, the enzyme reacts with a phospholipid to generate a phosphate group and an intermediate compound (Scheme 2, reaction 1). This later reacts with a molecule of water available in the reaction medium or blood medium of the patient (Scheme 2, reaction 2). These chemical transformations result in the regeneration of the enzyme or RNA polymerase as well as the decomposition of phospholipids (Scheme 2, reaction 2).
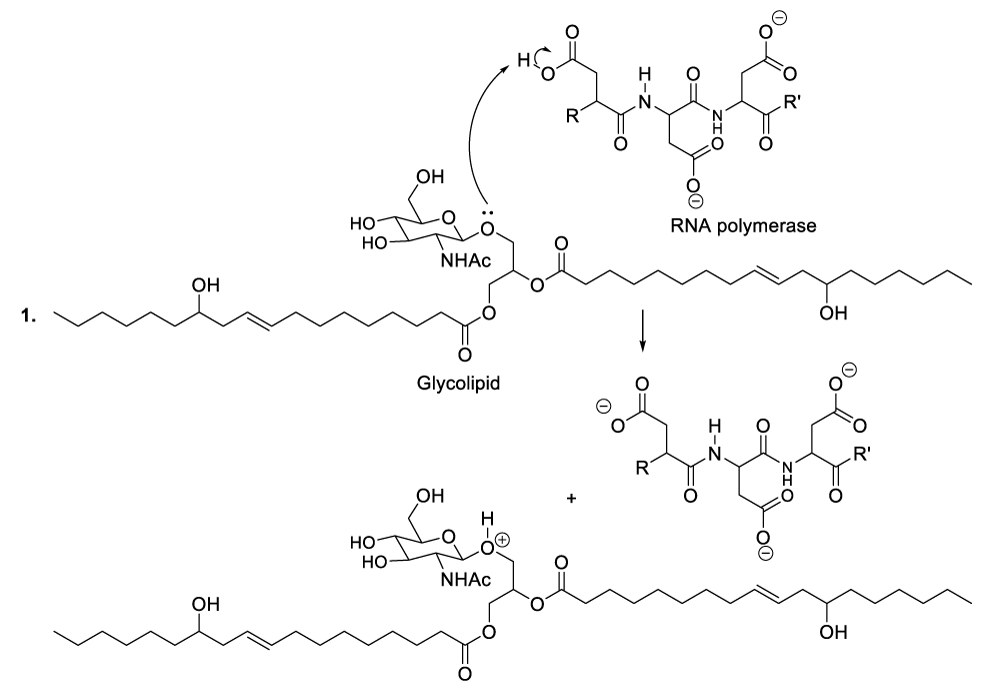
Enzymatic hydrolysis of glycolipids
The plausible reaction mechanism of the hydrolysis of glycolipids is accelerated by the RNA polymerase (Scheme 3, reaction 1). Indeed, the starting step of the hydrolysis of glycolipids is the glycolipid protonation via the reaction of glycolipid with the RNA polymerase. This step is followed by the inversion of the chair-like conformation of the protonated glycolipid to generate the twist-boat conformation in order to line up a lone pair of intra cyclic oxygen electrons into an antiperiplanar fashion with respect to the living group. This kind of living group, it will easily departure due to the electronic pressure because of the lone pair of intra cyclic oxygen electrons. This step finalizes the decomposition of the glycolipid since it permits the formation of the cyclic oxocarbonium ion and the lipid degradation moisture (Scheme 3, reaction 2). The following step involves the nucleophilic addition of the RNA polymerase conjugate base to the cyclic oxocarbonium ion resulting therefore to the esterification of the carbohydrate moisture, and this addition reaction allows the production of the corresponding compound (Scheme 3, reaction 3). The following steps demonstrate the action of a molecule of water to regenerate the RNA polymerase and the formation of the corresponding products (Scheme 3, reactions 4-7).
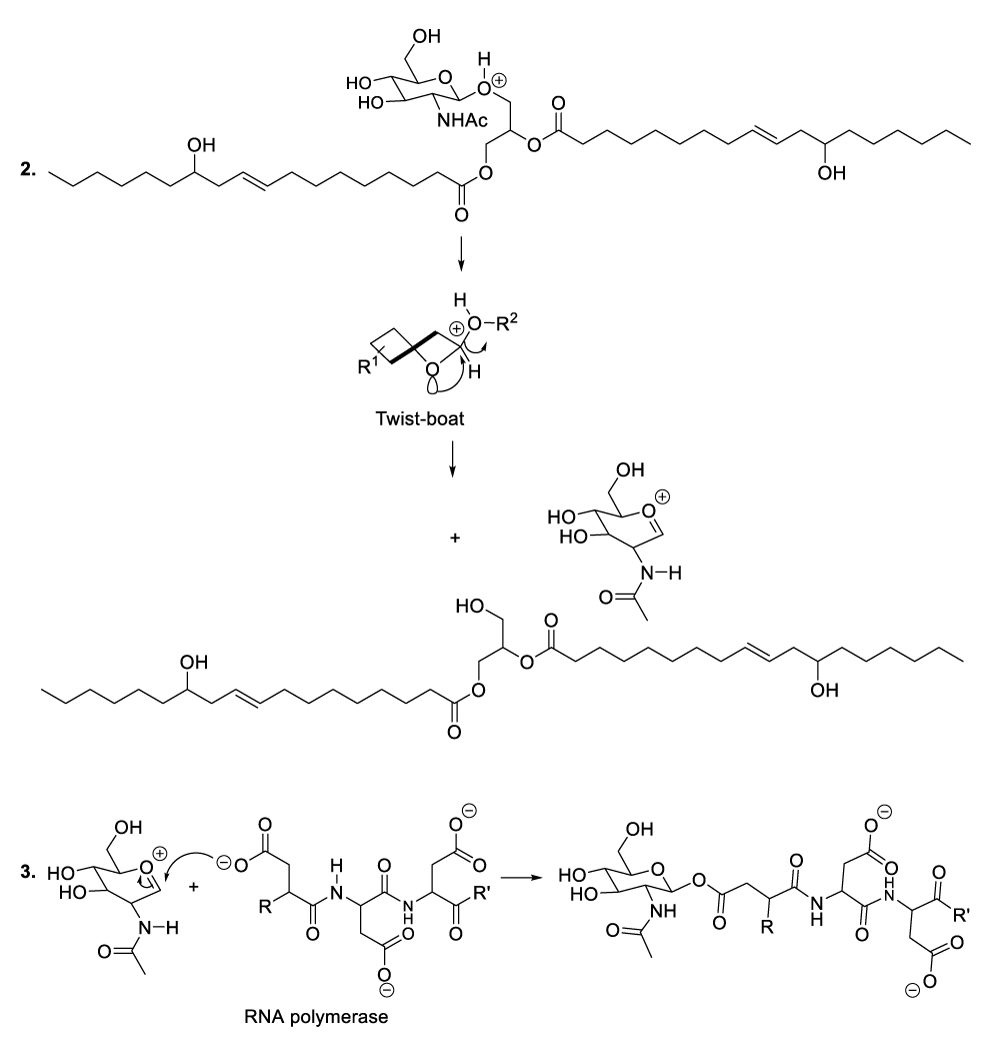
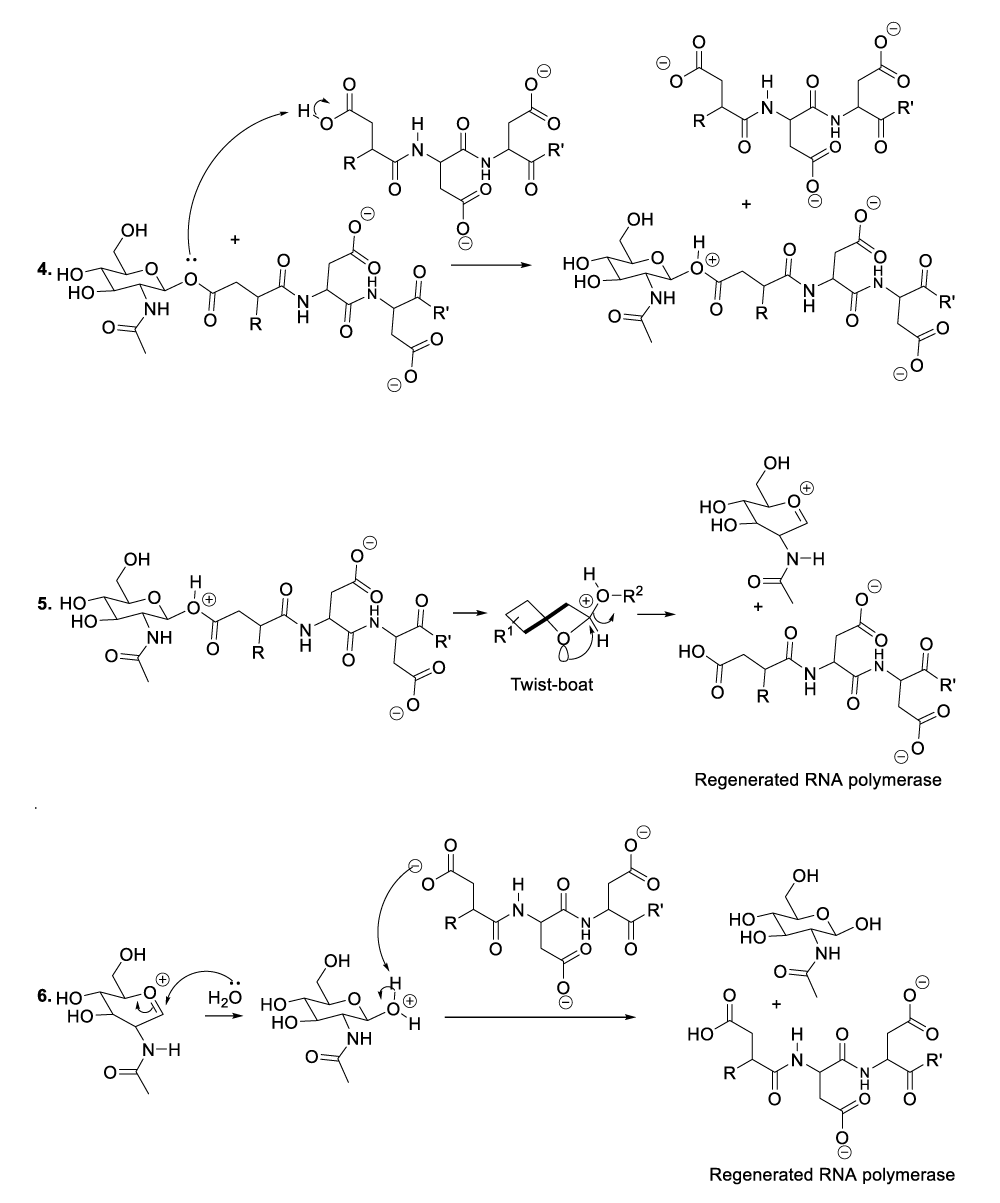
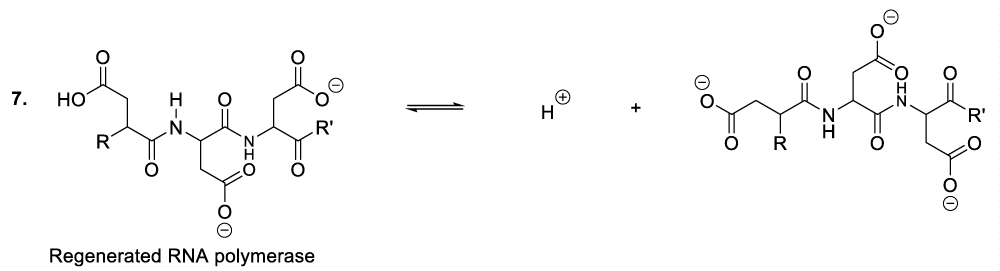
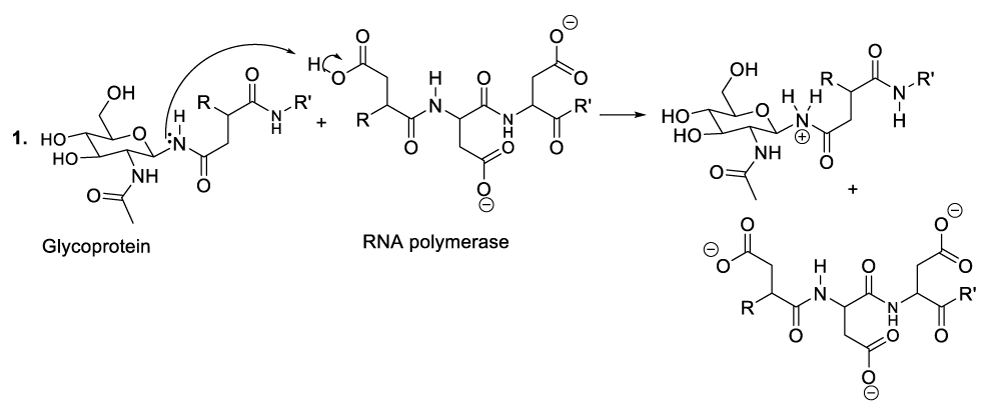
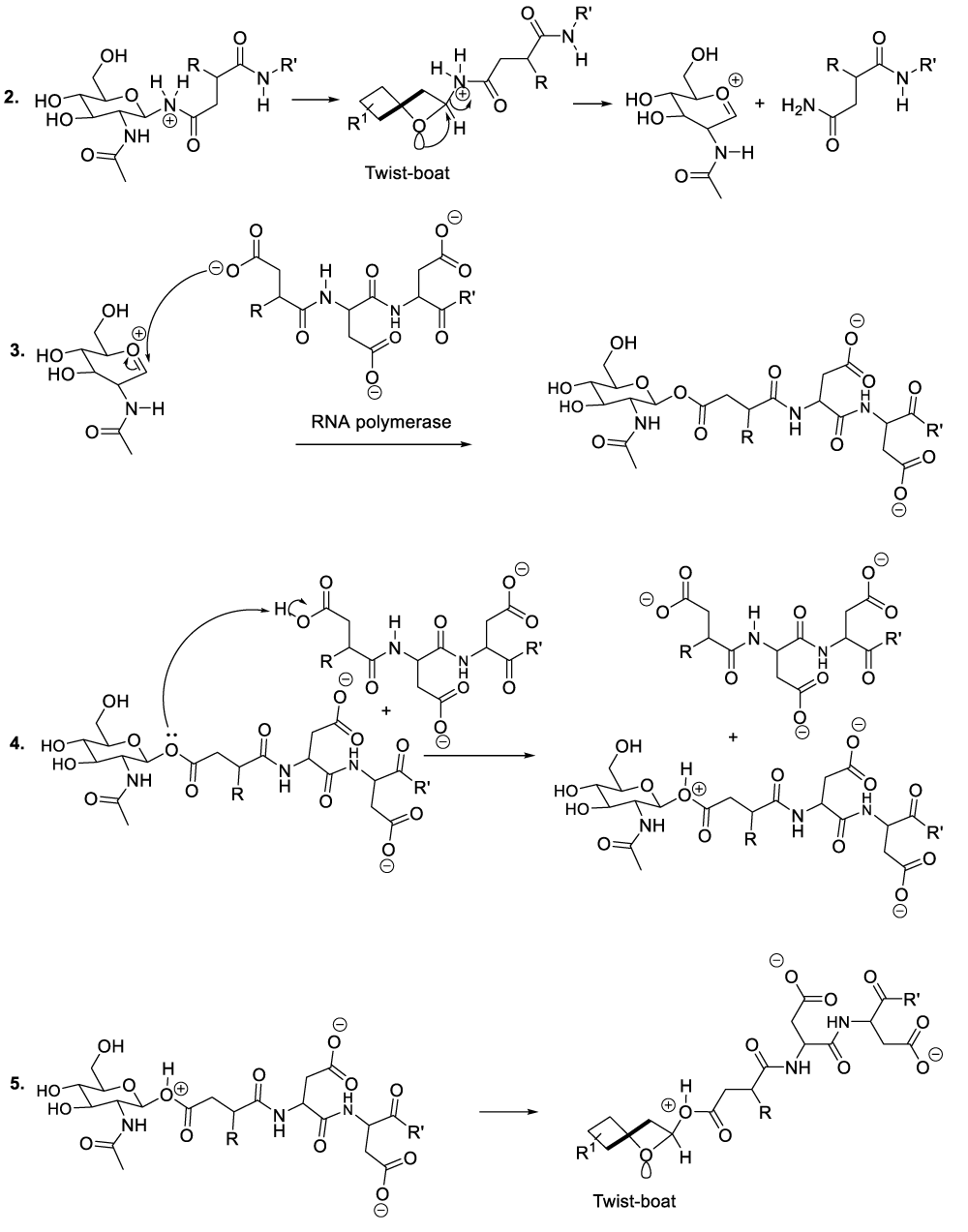
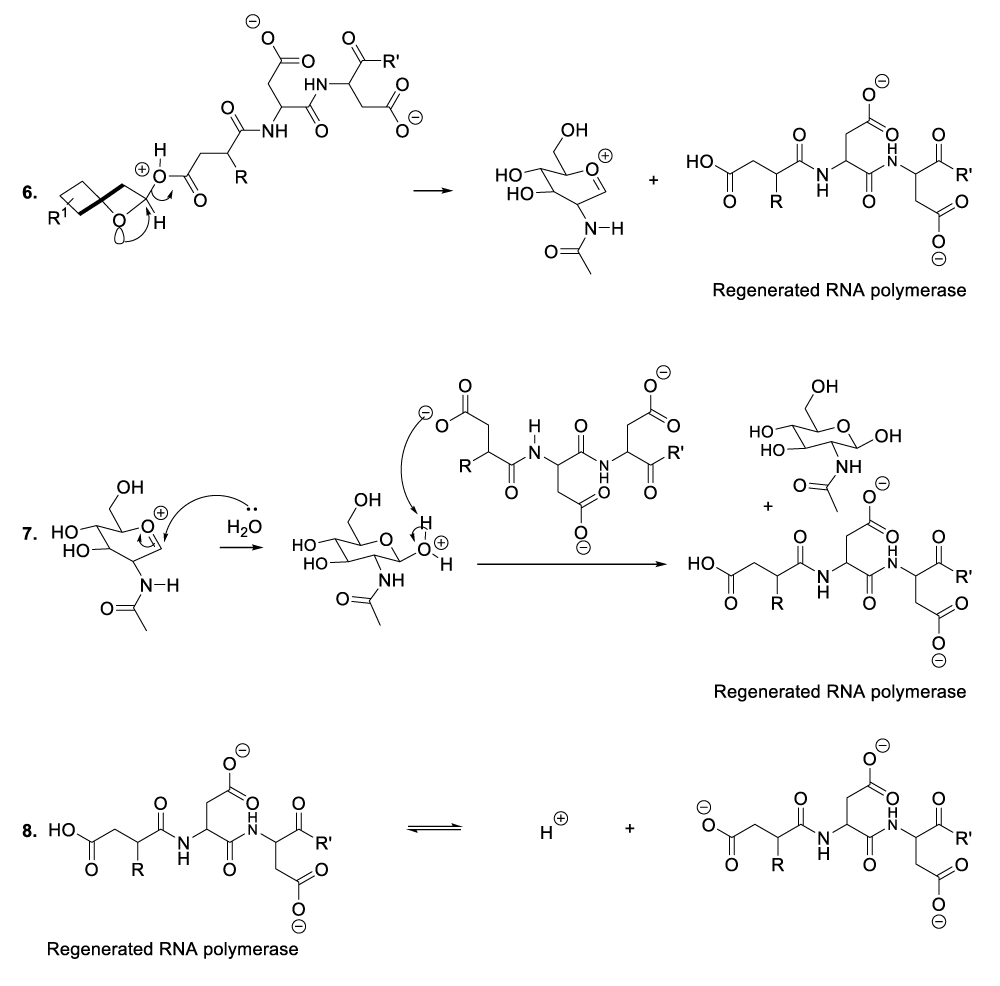
Enzymatic hydrolysis of glycoproteins
Hydrolysis of glycoproteins begins with the reaction between RNA polymerase and the appropriate glycoprotein to furnish the predicted intermediate product (Scheme 4, reaction 1).11 This step is followed by the inversion of the intermediate product chair-like conformation to adopt a twist-boat configuration in order to line up an intra-cyclic oxygen lone pair in an antiperiplanar manner to the living group in order to generate the oxocarbonium ion as well as the glycoprotein disintegration product or the protein fragment (Scheme 4, reaction 2). This part of the hydrolysis of glycoprotein is followed by the nucleophilic addition of the RNA polymerase to the oxocarbonium ion to furnish the adequate intermediate product (Scheme 4, reaction 3). The following stages finalize the glycoprotein hydrolysis reaction (Scheme 4, reactions 4-8).
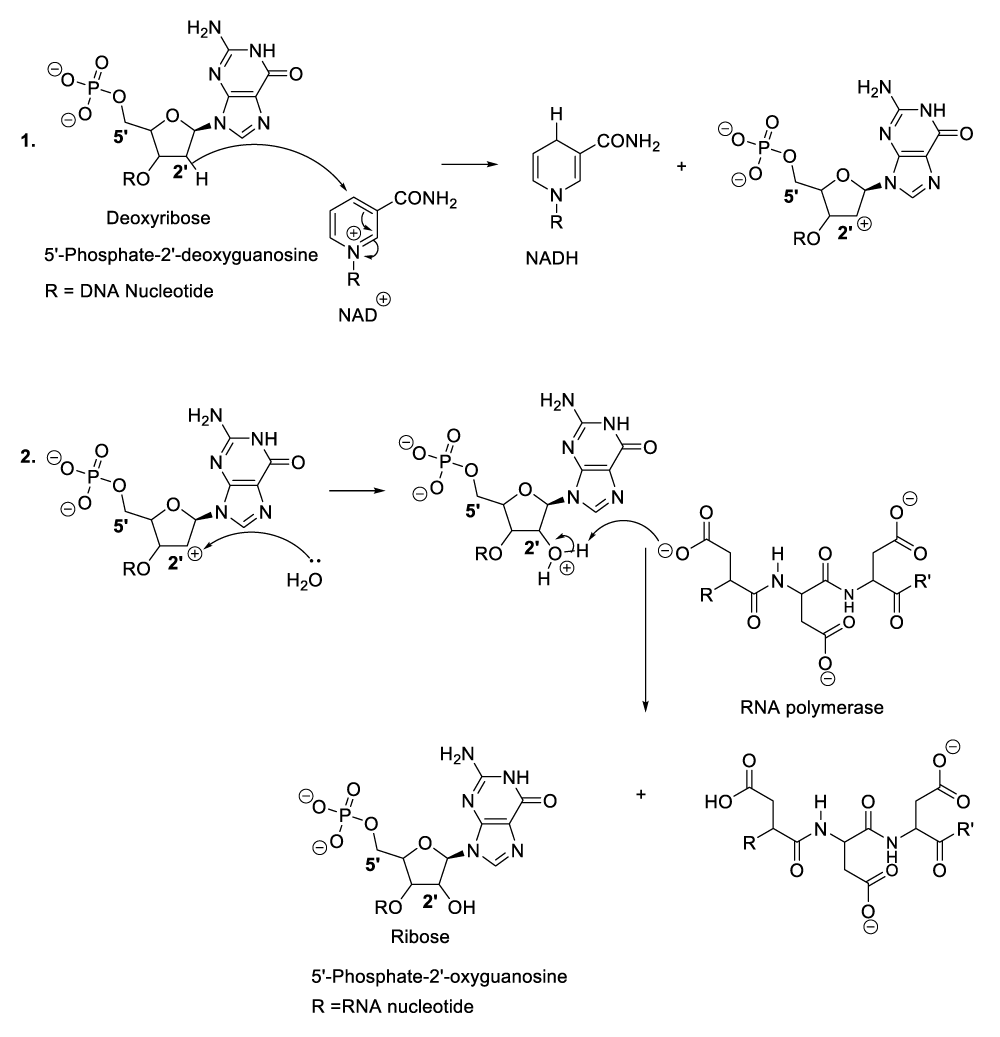
Enzymatic synthesis of RNA
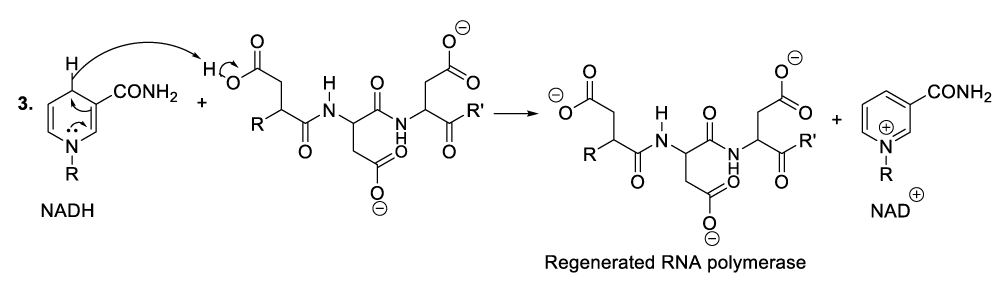
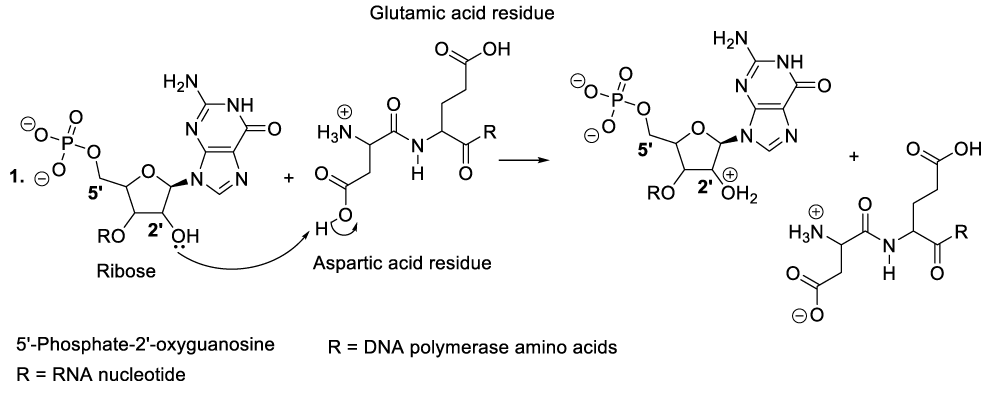
When bacteriophages penetrate the targeted bacteria, they spread or multiply and consequently, the excessive accumulation of bacteriophages into the bacterial cytoplasm leads to the degradation of the infectious bacteria. In order to achieve that, the bacteriophages synthesize many molecules of RNA by hydroxylation of DNA substrate (Scheme 5). The reasonable mechanism of his kind of organic synthesis starts with the nucleophilic addition of aspartate residue to the DNA carbon 2’ in order to eject a hydride ion, which will be trapped with a coenzyme or an oxidized molecule of nicotinamide adenine dinucleotide (NAD+) to provide the corresponding products (Scheme 5, reaction 1). This reaction sequence is followed by the hydroxylation of the intermediate product carbon 2’ with a molecule of water (Scheme 5, reaction 2). As a result, another intermediate product is formed of which the proton reacts with nicotinamide adenine dinucleotide hydride (NADH), and the regeneration of RNA polymerase as well as the formation of the oxidized state of NADH or nicotinamide adenine dinucleotide (NAD+) (Scheme 5, reaction 2, reaction 3).
Enzymatic synthesis of DNA
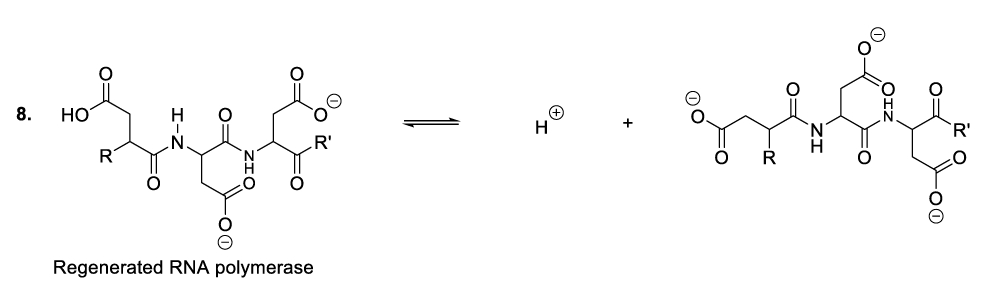
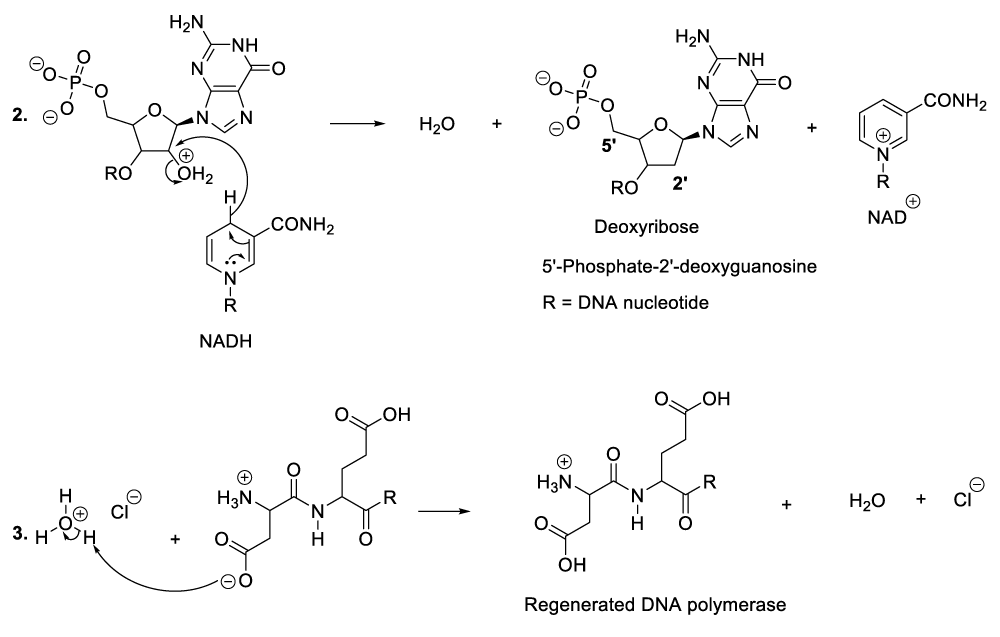
Within the bacterial cellular cytoplasm or bacterial intracellular reaction medium, bacteriophages also synthesize many molecules of DNA by elimination of an RNA hydroxide leaving group. This chemical conversion is catalyzed by aspartic and glutamic acid residues situated in the DNA polymerase active centre (Scheme 6) [12-19]. In this context, aspartic acid donates un proton to the carbon 2’ hydroxide group of a bacteriophage RNA to produce the corresponding compounds (Scheme 6, reaction 1). This reaction step is followed by the displacement of a leaving group (water molecule) due to the nucleophilic addition of a hydride ion from NADH. As a result, DNA molecules are formed as well as NAD+ including DNA polymerase (Scheme 6, reaction 2, reaction 3).
It is important to mention that aspartic and glutamic acids are essential amino acids for DNA polymerase catalytic activities. In the same perspective, chemical constituents such as NADH along with hydrochloric acid available in the cytosol (a liquid portion of cellular cytoplasm) play a fundamental role in the microorganism metabolic process [12-19].
Conclusion
I have shown the noteworthiness of the chemistry or reactivity of organic compounds to better understand bacteriophage therapy fashion. From this perspective, this category of medical approach has been demonstrated, in detail, taking advantage of the enzymatic organic reaction mechanisms. In other words, this conceptual study has revealed that bacteriophages hydrolyze bacterial cellular membrane phospholipids, glycolipids, and glycoproteins before reaching the bacterial cytoplasm where they synthesize ribonucleic acids as well as deoxyribonucleic acids, for the purpose of becoming preponderant to detriment of the pathogen bacteria. This bacteriophage pathway contributes enormously to the neutralization of the targeted bacteria.
Essentially, the enzymatic reactions exhibited in this article are reasonable in organic chemistry because usually, it is known that organic reactions take place upon functional groups of reactive entities. That is to say, functional groups are the root of organic compound reactivity. In that perspective, the enzymatic reactions developed in this conceptual research are a consequential added value, which can be applied to explaining the mechanism of action regarding therapeutic organic substances. It can be especially applied to demonstrate the chemistry or the reactivity of potentially anticancer organic compounds with respect to our ongoing research.
- Gao Z, Feng Y. Bacteriophage strategies for overcoming host antiviral immunity. Front Microbiol. 2023;14:1211793. Available from: https://doi.org/10.3389/fmicb.2023.1211793
- Labrie SJ, Samson JE, Moineau S. Bacteriophage resistance mechanisms. Nat Rev Microbiol. 2010;8(5):317-27. Available from: https://doi.org/10.1038/nrmicro2315
- Egido JE, Costa AR, Aparicio-Maldonado C, Haas PJ, Brouns SJJ. Mechanisms and clinical importance of bacteriophage resistance. FEMS Microbiol Rev. 2022;46(1):fuab048. Available from: https://doi.org/10.1093/femsre/fuab048
- Strathdee SA, Hatfull GF, Mutalik VK, Schooley RT. Phage therapy: From biological mechanisms to future directions. Cell. 2023;186(1):17-31. Available from: https://doi.org/10.1016/j.cell.2022.11.017
- Smith NM, Nguyen TD, Chin WH, Sanborn JT, de Souza H, Ho BM, Luong T, Roach DR. A mechanism-based pathway toward administering highly active N-phage cocktails. Front Microbiol. 2023;14:1292618. Available from: https://doi.org/10.3389/fmicb.2023.1292618
- Bleriot I, Pacios O, Blasco L, Fernández-García L, López M, Ortiz-Cartagena C, Barrio-Pujante A, García-Contreras R, Pirnay JP, Wood TK, Tomás M. Improving phage therapy by evasion of phage resistance mechanisms. JAC Antimicrob Resist. 2024;6(1):dlae017. Available from: https://doi.org/10.1093/jacamr/dlae017
- Callanan J, Stockdale SR, Shkoporov A, Draper LA, Ross RP, Hill C. RNA Phage Biology in a Metagenomic Era. Viruses. 2018;10(7):386. Available from: https://doi.org/10.3390/v10070386
- Tars, K. SsRNA phages: Life cycle, structure and applications Biocommunication of Phages 2020; 261-292. Available from: https://doi.org/10.1007/978-3-030-45885-0_13
- Laguna-Castro M, Rodríguez-Moreno A, Llorente E, Lázaro E. The balance between fitness advantages and costs drives adaptation of bacteriophage Qβ to changes in host density at different temperatures. Front Microbiol. 2023;14:1197085. Available from: https://doi.org/10.3389/fmicb.2023.1197085
- Nudler E. RNA polymerase active center: the molecular engine of transcription. Annu Rev Biochem. 2009;78:335-61. Available from: https://doi.org/10.1146/annurev.biochem.76.052705.164655
- Mwene-Mbeja TM, Dufour A, Lecka J, Kaur BS, Vaneeckhaute C. Enzymatic reactions in the production of biomethane from organic waste. Enzyme Microb Technol. 2020;132:109410. Available from: https://doi.org/10.1016/j.enzmictec.2019.109410
- Bressanelli S, Tomei L, Roussel A, Incitti I, Vitale RL, Mathieu M, De Francesco R, Rey FA. Crystal structure of the RNA-dependent RNA polymerase of hepatitis C virus. Proc Natl Acad Sci U S A. 1999;96(23):13034-9. Available from: https://doi.org/10.1073/pnas.96.23.13034
- Patel PH, Loeb LA. DNA polymerase active site is highly mutable: evolutionary consequences. Proc Natl Acad Sci U S A. 2000;97(10):5095-100. Available from: https://doi.org/10.1073/pnas.97.10.5095
- Batra VK, Perera L, Lin P, Shock DD, Beard WA, Pedersen LC, Pedersen LG, Wilson SH. Amino acid substitution in the active site of DNA polymerase β explains the energy barrier of the nucleotidyl transfer reaction. J Am Chem Soc. 2013;135(21):8078-88. Available from: https://doi.org/10.1021/ja403842j
- Wu CH, Haja DK, Adams MWW. Cytoplasmic and membrane-bound hydrogenases from Pyrococcus furiosus. Methods Enzymol. 2018;613:153-168. Available from: https://doi.org/10.1016/bs.mie.2018.10.009
- Schäfer C, Bommer M, Hennig SE, Jeoung JH, Dobbek H, Lenz O. Structure of an Actinobacterial-Type [NiFe]-Hydrogenase Reveals Insight into O2-Tolerant H2 Oxidation. Structure. 2016;24(2):285-92. Available from: https://doi.org/10.1016/j.str.2015.11.010
- Chandrayan SK, McTernan PM, Hopkins RC, Sun J, Jenney FE Jr, Adams MW. Engineering hyperthermophilic archaeon Pyrococcus furiosus to overproduce its cytoplasmic [NiFe]-hydrogenase. J Biol Chem. 2012;287(5):3257-64. Available from: https://doi.org/10.1074/jbc.m111.290916
- Lewis CA, Parker SJ, Fiske BP, McCloskey D, Gui DY, Green CR, Vokes NI, Feist AM, Vander Heiden MG, Metallo CM. Tracing compartmentalized NADPH metabolism in the cytosol and mitochondria of mammalian cells. Mol Cell. 2014;55(2):253-63. Available from: https://doi.org/10.1016/j.molcel.2014.05.008
- Venning JD, Grimley RL, Bizouarn T, Cotton NP, Jackson JB. Evidence that the transfer of hydride ion equivalents between nucleotides by proton-translocating transhydrogenase is direct. J Biol Chem. 1997;272(44):27535-8. Available from: https://doi.org/10.1074/jbc.272.44.27535
Article Alerts
Subscribe to our articles alerts and stay tuned.
 This work is licensed under a Creative Commons Attribution 4.0 International License.
This work is licensed under a Creative Commons Attribution 4.0 International License.


 Save to Mendeley
Save to Mendeley
