Open Journal of Chemistry
Potential of Imiquimod and Fulvestrant against Mycobacterium tuberculosis using Molecular docking approach
Asma Noor*, Muhammad Imran Qadir, Ardas Masood, Jawaria Khan and Sania Ahmad
Cite this as
Noor A, Qadir MI, Masood A, Khan J, Ahmad S (2017) Potential of Imiquimod and Fulvestrant against Mycobacterium tuberculosis using Molecular docking approach. Peertechz J Med Chem Res 3(1): 023-024. DOI: 10.17352/ojc.000008Mycobacterium Tuberculosis cause severe disease of lungs known as Tuberculosis. It is a major cause of morbidity and mortality even in the emerging countries also. However, to prepare an antibiotics drug against Mycobacterium tuberculosis is a major challenge. Two compounds Imiquimod, Fulvestrant were selected for our study which can stop action of Arbinosyl transferase enzyme of M.tuberculosis. In silico docking was carried out by using software named swissdock (https://www.swissdock.ch/). These are computer aided drug designing techniques which are helpful for this study. Both the above 2 drugs have the potential to block the activity of mutant strain of M.tuberculosis.
Introduction
Tuberculosis is a disease of lungs cause by bacteria Mycobacterium tuberculosis. This bacterium is found in lungs but if it is not treated it can be spread to other parts of body. About 33% of population of world is considered to be infected through this virus. A sick person can also transfer this bacterium to healthy persons through mouth or air [1-3]. Antibiotics are mostly used to treat this infectious disease. Effective antibiotics target the cell wall of bacteria and stop their multiplication in lungs. It usually takes place 6-9 months to treat this disease.
Arabinosyl transferase enzyme in M.tuberclosis is the cause of Tuberculosis in human being. The docking results show that above selected ligands have the ability to bind with the cell wall of bacteria and cell wall permeability also increases which causes the death of bacteria.
TB Diagnosis relies on skin test of tuberculosis or (TST) or tests of blood. Although its treatment is also hard and involves disposal of several antibiotics for the long interval of time. Social links also screened or handled if necessary. The resistance of antibiotics is also an emerging problem in multiple or several drug-resistant tuberculosis (MDR-TB) infections. Currently a large variety of novel vaccines are useable in the development but it is also a major killing bacteria for developing countries.
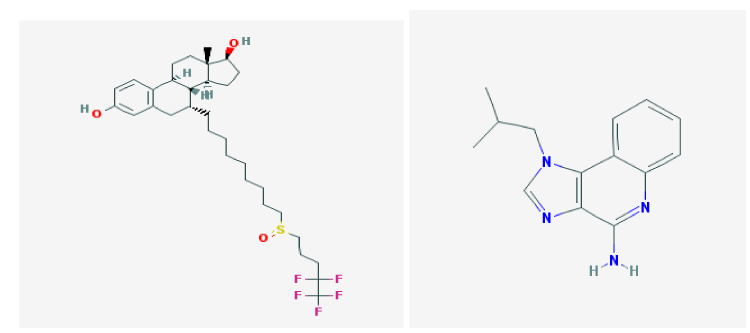
Fulvestrant is first selected drug use in our study. It also acts as an anti-cancerous compound by blocking the abnormal hormone production in the body. It also acts as an anti-cancerous compound, antioxidant, anti-inflammatory [4]. However adverse effects include head ache, bone pain, sore throat, weakness, nausea and vomiting.
Imiquimod acts as immune system modifier. It also acts as an anti-cancerous compound, antioxidant, anti-inflammatory, anti -allergic, prohibition of various enzymes have been inquired [5]. (However, adverse consequences are headaches, pain in back, aches of muscles, fatigue, diarrhea, and fungal infections).
Material and Methods
Receptor and ligand retrieval and analogs design of analog for Imiquimod and Fulvestrant
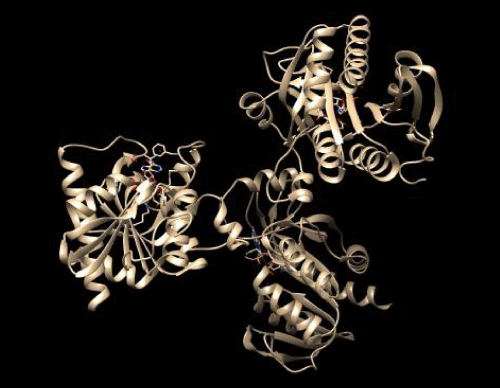
Arbinosyl transferase (3PTY) 3-D structure was recollected from data bank of protein (pdb) as (Figure 1). Similarly the structure for imiquimod and Fulvestrant were obtained also from Drug Bank .Similarity of structure, sub-structure, identification (70%) search performed and behaved out of these chemical compounds by using Browser Molsoft ICM 3.5-1p and software name Chem BioDraw Ultra 12.0. Compound library was collected from ZINC Database, PubChem.
Optimization of Ligand structure and calculation of physiochemical properties
The optimization of screened ligands were done before by using force field MM2 of Chem Bio 3D ultra[6]. The structure of ligands like Imiquimod and Fulvestrant, were downloaded from Pubchem (https://www.ncbi.nlm.nih.gov/pccompound) and stored in UCSF chimera 1.10.1 version of software.
Result and Discussion
The physiochemical properties (Hydrogen bond acceptor, hydrogen bond donor, number of rotatable bond, Calculated, molecular weight, etc) were predicted and checked for non-violation of drug all are given below. Moreover the 2D structures of selected ligands are also shown in figure 2.
Potential protein binding sites prediction and molecular docking study
The potential ligand binding site of Arbinosyl transferase receptor was computed at Swiss dock. Volume and Surface of the binding site were computed and optimum binding site was selected to perform[7]. The screened out compounds were spelt or imported in Swiss docking software. Swiss dock software is use for visualization and docking at molecular level[8]. The flexibility of bonds in ligands were lay down and similarly the flexibility of side chain of amino acids into the fastening or binding cavity was also adjust on a tolerance of 1.10 ,strength of 0.90 towards docking simulations. Fundamental interaction of Ligands for receptor was considered to determine the better binding conformation of complex of receptor-ligand. The scoring for imiquimod and Fulvestrant are 12 and 15 respectively.
Medcam designer software is used to ADMET our ligand. ADMET properties of the compounds Fulvestrant, imiquimod are tabulated respectively in the table 1.
ADMET and prediction of toxicity
Absorption, Distribution, Metabolism, Excretion and Toxicity all the properties were studied for above mentioned compounds and were computed by using PASS (Prediction of Activity Spectra for Substances) Inet and Pre ADMET server). PASS and Inet predicts 2536 of 4130 possible activities as immune stimulant, kidney function stimulator, fibronylitic, antiviral, anti-leukemic etc were studied [9]. However, some adverse effects are cardio toxicity, hypo-calcemic, depression, nephro-toxic were studied.
It is a serious disease and it is necessary to develop a new drug for Mycobacterium tuberculosis. In this study we examined many compounds but two anticancer compounds were selected for further studies on the basis of minimum score and top conformation.
Docking score and H-bond interaction of ligands against Mycobacterium tuberculosis is given table 2.
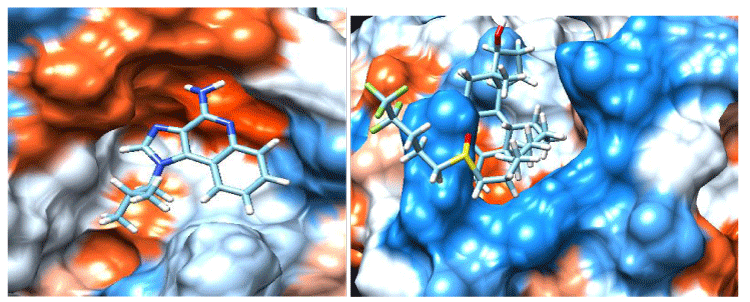
Docked poses of ligand Imiquimod, Fulvestrant with the enzyme shown in figure 3.
After successful docking of the Fulvestrant and Imiquimod with Arbinosyl transferase the complex models of receptor/ligand was retrived and this analysis depends on the parameter such as interaction of hydrogen bond, interactions ï – ï, energy of binding.
Conclusion
Molecular docking belongs to a new innovative approach for the study of small compound binding with the receptor protein. The suitable ligand is that whose hydrogen bond donor is less than five. The ligand we selected for study form hydrogen bond with the catalytic triad of protein. The ligand we selected has potential against mycobacterium tuberculosis
- Amin AG, Goude R, Shi L, Zhang J, Chatterjee D et al. (2008) EmbA is an essential arabinosyltransferase in Mycobacterium tuberculosis. J Microbiology 154, 240-248. Link: https://goo.gl/pWW3Ky
- Zoete V, A. Grosdidier and O. Michielin (2009) Docking, virtual high throughput screening and in silico fragment́‐based drug design. Journal of cellular and molecular medicine 13: 238-248. Link: https://goo.gl/5ebxyJ
- McIlleron H, Willemse M, Werely CJ, Hussey GD, Schaaf HS et al. (2009) Isoniazid plasma concentrations in a cohort of South African children with tuberculosis: Implications for international pediatric dosing guidelines. J Clin Infect Dis.48: 1547-53. Link: https://goo.gl/PF6fkB
- Osborne C, J. Pippen, S.E. Jones, L.M. Parker, M. Ellis et al. (2002) Double-blind, randomized trial comparing the efficacy and tolerability of fulvestrant versus anastrozole in postmenopausal women with advanced breast cancer progressing on prior endocrine therapy: results of a North American trial. Journal of Clinical Oncology, 20: 3386-3395. Link: https://goo.gl/788gaJ
- Lebwohl M, ScottDinehart, DavidWhiting, Peter KLee, NajiTawfik et al. (2004) Imiquimod 5% cream for the treatment of actinic keratosis: results from two phase III, randomized, double-blind, parallel group, vehicle-controlled trials. Journal of the American Academy of Dermatology, 50: 714-721. Link: https://goo.gl/opypBd
- Thai K.-M and G.F. Ecker. (2007) Predictive models for HERG channel blockers: ligand-based and structure-based approaches. Current medicinal chemistry,14: 3003-3026. Link: https://goo.gl/wZSW3x
- Kolita B, Dhrubajyoti Gogoi, Partha Pratim Dutta, Manobjyoti Bordoloi, Rajib Lochan Bezbaruah et al. (2014) Arabinosyl transferase inhibitor design against Mycobacterium tuberculosis using ligand based drug design approach. Bangladesh Journal of Pharmacology, 9: 225-229. Link: https://goo.gl/uAqxxa
- Grosdidier A, V. Zoete, O. Michielin and SwissDock. (2011) a protein-small molecule docking web service based on EADock DSS. Nucleic acids research, 39: 270-277. Link: https://goo.gl/k8trbT
- Campillo N.E. and J.A. (2009) Páez, In Silico ADME Approaches. Frontiers in Drug Design and Discovery, 1: 291.
Article Alerts
Subscribe to our articles alerts and stay tuned.
 This work is licensed under a Creative Commons Attribution 4.0 International License.
This work is licensed under a Creative Commons Attribution 4.0 International License.

 Save to Mendeley
Save to Mendeley
